For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELDNDPGIGAIIITGSAKAFAAGADIKEM ADLTFSEAFDADFFAAWGKLAAVRTPTIAAVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLPGM GGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKAVATTISQMSRSAARMAKEA VNRTFEATLAEGLLYERRLIHSAFATADQSEGMAAFIEKRPPNFTHR
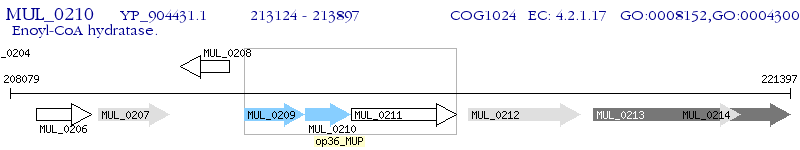
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0210 | echA8 | - | 100% (257) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_4076 | echA19 | 2e-35 | 36.33% (256) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_1899 | echA17 | 3e-35 | 36.97% (238) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 1e-129 | 91.05% (257) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 1e-117 | 81.32% (257) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 1e-129 | 91.05% (257) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-123 | 86.77% (257) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 1e-113 | 79.69% (256) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4396 | echA8 | 1e-140 | 100.00% (257) | enoyl-CoA hydratase EchA8 |
| M. avium 104 | MAV_1195 | - | 1e-126 | 88.67% (256) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5277 | - | 1e-114 | 81.57% (255) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_4102 | - | 1e-119 | 84.38% (256) | PUTATIVE short chain enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4681 | - | 1e-113 | 80.08% (256) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2036|M.gilvum_PYR-GCK ------MTFETILVDRDDRVATITLNRPKALNALNTQVMNEVTTAAAELD
Mvan_4681|M.vanbaalenii_PYR-1 ---MGSQNFETIIVERDDRVATITLNRPKALNALNSQVMDEVTTAAAELD
MSMEG_5277|M.smegmatis_MC2_155 -----MSDFETILVTRTDRVATIQLNRPKALNALNSQVMTEVTAAAAELD
TH_4102|M.thermoresistible__bu ---MSAKNYETILVTREDRVGIITLNRPKALNALNSQVMTEVTDAAAEFD
Mb1099c|M.bovis_AF2122/97 ------MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELD
Rv1070c|M.tuberculosis_H37Rv ------MTYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAATELD
MUL_0210|M.ulcerans_Agy99 ------MSYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELD
MMAR_4396|M.marinum_M ------MSYETILVERDQRVGIITLNRPQALNALNSQVMNEVTSAAAELD
MLBr_02402|M.leprae_Br4923 ------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAAKELD
MAV_1195|M.avium_104 MTDSTDRTFETILVEREERVGIITLNRPKALNALNTQVMNEVTTAATDFD
MAB_1185c|M.abscessus_ATCC_199 ---MTSHNFETILTERIDRVAVITLNRPKALNALNSQVMNEVTTAAAEFD
::**:. :**. * ****:******:*:* *:* ** ::*
Mflv_2036|M.gilvum_PYR-GCK AEPGVGAIIVTGSERAFAAGADIKEMASLSFADVFSSDFFAAWGKFAATR
Mvan_4681|M.vanbaalenii_PYR-1 ADAGVGAIILTGSEKAFAAGADIKEMAGLSFADVFSSDFFAGWGKFAATR
MSMEG_5277|M.smegmatis_MC2_155 KDPGVGAIIVTGNEKAFAAGADIKEMADLSFAEVFEADFFELWSKFAATR
TH_4102|M.thermoresistible__bu SDPGIGAIIVTGNEKAFAAGADIKEMADLEFADVFAADFFAPWSKFAATR
Mb1099c|M.bovis_AF2122/97 DDPDIGAIIITGSAKAFAAGADIKEMADLTFADAFTADFFATWGKLAAVR
Rv1070c|M.tuberculosis_H37Rv DDPDIGAIIITGSAKAFAAGADIKEMADLTFADAFTADFFATWGKLAAVR
MUL_0210|M.ulcerans_Agy99 NDPGIGAIIITGSAKAFAAGADIKEMADLTFSEAFDADFFAAWGKLAAVR
MMAR_4396|M.marinum_M NDPGIGAIIITGSAKAFAAGADIKEMADLTFSEAFDADFFAAWGKLAAVR
MLBr_02402|M.leprae_Br4923 IDPDVGAILITGSPKVFAAGADIKEMASLTFTDAFDADFFSAWGKLAAVR
MAV_1195|M.avium_104 ADPGIGAIIITGSAKAFAAGADIKEMAELTFADAYGADFFAPWGKLAALR
MAB_1185c|M.abscessus_ATCC_199 ADHGIGAIIITGSEKAFAAGADIKEMSEQSFSDMFGSDFFSAWGKLGAVR
: .:***::**. :.**********: *:: : :*** *.*:.* *
Mflv_2036|M.gilvum_PYR-GCK TPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQ
Mvan_4681|M.vanbaalenii_PYR-1 TPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQ
MSMEG_5277|M.smegmatis_MC2_155 TPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGSQ
TH_4102|M.thermoresistible__bu TPTIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQ
Mb1099c|M.bovis_AF2122/97 TPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
Rv1070c|M.tuberculosis_H37Rv TPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
MUL_0210|M.ulcerans_Agy99 TPTIAAVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLPGMGGSQ
MMAR_4396|M.marinum_M TPTIAAVAGYALGGGCEVAMMCDLIIAADTAKFGQPEIKLGVLPGMGGSQ
MLBr_02402|M.leprae_Br4923 TPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQ
MAV_1195|M.avium_104 TPTIAAVAGHALGGGCELAMMCDVLIAADTAKFGQPEIKLGVLPGMGGSQ
MAB_1185c|M.abscessus_ATCC_199 TPTIAAVSGYALGGGCELAMMCDVIIAAENATFGQPEIKLGVLPGMGGSQ
** ****:*:*******:*****::***:.*.******************
Mflv_2036|M.gilvum_PYR-GCK RLTRAIGKAKAMDLILTGRNMDAEEAERAGLVSRVVPADSLLSEAKAVAQ
Mvan_4681|M.vanbaalenii_PYR-1 RLTRAIGKAKAMDLILTGRNMGAEEAERAGLVSRVVPADTLLEEARSVAK
MSMEG_5277|M.smegmatis_MC2_155 RLTRAIGKAKAMDLILTGRTIDAAEAERAGLVSRLVPADTLIDEALAVAQ
TH_4102|M.thermoresistible__bu RLTRAIGKAKAMDLILTGRTIDAEEAERSGLVSRVVPADKLLDEAKETAA
Mb1099c|M.bovis_AF2122/97 RLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTEARATAT
Rv1070c|M.tuberculosis_H37Rv RLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVVPADDLLTEARATAT
MUL_0210|M.ulcerans_Agy99 RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKAVAT
MMAR_4396|M.marinum_M RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKAVAT
MLBr_02402|M.leprae_Br4923 RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVAT
MAV_1195|M.avium_104 RLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVPADDLLTEAKKVAT
MAB_1185c|M.abscessus_ATCC_199 RLTRAIGKAKAMDMILTGRNMDAAEAERSGLVSRVVATESLLDEAKAVAK
*************:*****.:.* ****:*****:* :: *: ** .*
Mflv_2036|M.gilvum_PYR-GCK TIAGMSLSASRMAKEAVDRAFETTLSEGLLYERRLFHSAFATDDQTEGMN
Mvan_4681|M.vanbaalenii_PYR-1 TIAGMSLSASRMAKEAVDRAFETTLTEGLLYERRLFHSAFATDDQTEGMN
MSMEG_5277|M.smegmatis_MC2_155 TIAGMSLSASRMAKEAVNRAFETSLTEGLLYERRLFHSAFATADQKEGMA
TH_4102|M.thermoresistible__bu TIAAMSLSAARMAKEAVNRAFETSLAEGLLFERRVFHSAFATADQSEGMA
Mb1099c|M.bovis_AF2122/97 TISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMA
Rv1070c|M.tuberculosis_H37Rv TISQMSASAARMAKEAVNRAFESSLSEGLLYERRLFHSAFATEDQSEGMA
MUL_0210|M.ulcerans_Agy99 TISQMSRSAARMAKEAVNRTFEATLAEGLLYERRLIHSAFATADQSEGMA
MMAR_4396|M.marinum_M TISQMSRSAARMAKEAVNRTFEATLAEGLLYERRLIHSAFATADQSEGMA
MLBr_02402|M.leprae_Br4923 TISQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMA
MAV_1195|M.avium_104 TISQMSRSAARMAKEAVNRAFETTLAEGLLYERRLFHSTFATADQSEGMA
MAB_1185c|M.abscessus_ATCC_199 TISEMSLSASMMAKEAVNRAFESSLAEGLLFERRIFHSAFGTADQSEGMA
**: ** **: ******:*:**::*:****.***::**:* * **.***
Mflv_2036|M.gilvum_PYR-GCK AFSEKRSPNFTHR
Mvan_4681|M.vanbaalenii_PYR-1 AFTEKRSPNFTHR
MSMEG_5277|M.smegmatis_MC2_155 AFAEKRAANFTHR
TH_4102|M.thermoresistible__bu AFTEKRPPKFTHR
Mb1099c|M.bovis_AF2122/97 AFIEKRAPQFTHR
Rv1070c|M.tuberculosis_H37Rv AFIEKRAPQFTHR
MUL_0210|M.ulcerans_Agy99 AFIEKRPPNFTHR
MMAR_4396|M.marinum_M AFIEKRPPNFTHR
MLBr_02402|M.leprae_Br4923 AFIEKRAPQFTHR
MAV_1195|M.avium_104 AFVEKRAPNFTHR
MAB_1185c|M.abscessus_ATCC_199 AFVEKRPANFIHR
** ***..:* **