For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIVTVTPNPSLDRTLHLPALRPGEVNRATSTMTEPSGKGVNVALALHGAGHATRAVLPVGGSVGAEIASA LEGFGLDTIGVPVAGSVRSNVTLVEADGRSTKVNEPGPFLSEDEVDALCAAALADAADWVVWAGSLPAGF APARLAAAVDEARAAGRRVAVDASGAAMTAVLAGGSPHLIKPNLDELADATGATPATLADVVDAARSLRV DTVLVSLGGDGAVLVTSDLALHGTAPVDRVVNTVGAGDALLAGYLAACSDGPADALASALRFGATAVEHE GTLIGVPDPQRPVRIGPARGDLRLG
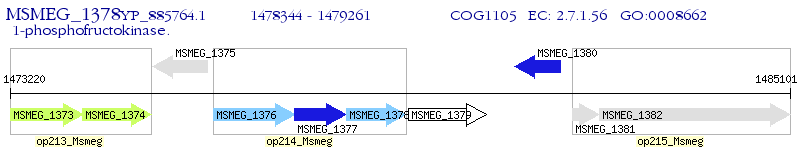
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1378 | - | - | 100% (305) | 1-phosphofructokinase |
| M. smegmatis MC2 155 | MSMEG_0086 | - | 9e-43 | 41.37% (307) | 1-phosphofructokinase |
| M. smegmatis MC2 155 | MSMEG_3947 | - | 1e-28 | 34.28% (283) | 6-phosphofructokinase isozyme 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2054c | pfkB | 4e-28 | 32.04% (284) | phosphofructokinase pfkB (phosphohexokinase) |
| M. gilvum PYR-GCK | Mflv_0751 | - | 2e-45 | 41.69% (307) | ribokinase-like domain-containing protein |
| M. tuberculosis H37Rv | Rv2029c | pfkB | 4e-28 | 32.04% (284) | phosphofructokinase PfkB (phosphohexokinase) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1617c | - | 3e-05 | 27.56% (283) | ribokinase RbsK |
| M. marinum M | MMAR_3482 | pfkB_1 | 7e-33 | 36.97% (284) | phosphofructokinase, PfkB_1 |
| M. avium 104 | MAV_1743 | - | 8e-13 | 31.29% (278) | kinase, pfkB family protein |
| M. thermoresistible (build 8) | TH_1728 | pfkB | 2e-29 | 33.94% (274) | Probable phosphofructokinase PfkB (PHOSPHOHEXOKINASE) |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0095 | - | 3e-42 | 40.07% (307) | ribokinase-like domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0751|M.gilvum_PYR-GCK ------------MIVTVTPNPSIDRTVTLPSALTRGAVHRVTSVTSQAGG
Mvan_0095|M.vanbaalenii_PYR-1 ------------MIVTVTANPSIDRTVSLPSPLTRGAVHRVTSVTNQAGG
MSMEG_1378|M.smegmatis_MC2_155 ------------MIVTVTPNPSLDRTLHLP-ALRPGEVNRATSTMTEPSG
Mb2054c|M.bovis_AF2122/97 MTEPAAWDEGKPRIITLTMNPALDITTSVD-VVRPTEKMRCGAPRYDPGG
Rv2029c|M.tuberculosis_H37Rv MTEPAAWDEGKPRIITLTMNPALDITTSVD-VVRPTEKMRCGAPRYDPGG
MMAR_3482|M.marinum_M MTSPADPAVRGPQIVTLTMNPALDITTCVG-VVRPTEKLRCSETRYDPGG
TH_1728|M.thermoresistible__bu ------------------MNPALDISTTTD-RVHHTSKVRCHGVRHDPGG
MAB_1617c|M.abscessus_ATCC_199 ----------------------MDLTLRVEELPAPGATVLATSAMPAPGG
MAV_1743|M.avium_104 --------------------MNLDLSLAVDALPRPGETVLASSLRQAPGG
:* : ..*
Mflv_0751|M.gilvum_PYR-GCK KGVNVAKVLTMAGVDTVAVLPA---AANDPLITALQAAAVPHRIVTTAEP
Mvan_0095|M.vanbaalenii_PYR-1 KGVNVAKVLTMAGVDAVAVLPA---AANDPLLHALQIAAVPYRAVTTTEP
MSMEG_1378|M.smegmatis_MC2_155 KGVNVALALHGAGHATRAVLPVG-GSVGAEIASALEGFGLDTIGVPVAGS
Mb2054c|M.bovis_AF2122/97 GGINVARIVHVLGGCSTALFPAG-GSTGSLLMALLGDAGVPFRVIPIAAS
Rv2029c|M.tuberculosis_H37Rv GGINVARIVHVLGGCSTALFPAG-GSTGSLLMALLGDAGVPFRVIPIAAS
MMAR_3482|M.marinum_M GGINVARIAHVLGGSVFAVFPVG-GSHGAHVSKLLSEAGVAFGEIPIAAS
TH_1728|M.thermoresistible__bu GGINVARIAHVLGAPVTAVFPIG-GHTGELLTDMLEHSGVSYHSVPIADL
MAB_1617c|M.abscessus_ATCC_199 KGANQAVAAARAGASVQFIGAVGSDGAAPMLRSHLADNNVGLDGLVEVDG
MAV_1743|M.avium_104 KGGNQAVAAARAGARVQFVGAVGDDAAAGQLRAHLQANGVGLDGAVEIPG
* * * * : . : * :
Mflv_0751|M.gilvum_PYR-GCK ARTNLTITEP-DGTTTKINEPGATLDDAALQAFSAAVLDASGNADWVVMS
Mvan_0095|M.vanbaalenii_PYR-1 ARTNLTITEP-DGTTTKINEPGAKLDAATLEAFTDAVLDAAGGADWVVMS
MSMEG_1378|M.smegmatis_MC2_155 VRSNVTLVEA-DGRSTKVNEPGPFLSEDEVDALCAAALADA--ADWVVWA
Mb2054c|M.bovis_AF2122/97 TRESFTVNESRTAKQYRFVLPGPSLTVAEQEQCLDELRGAAASAAFVVAS
Rv2029c|M.tuberculosis_H37Rv TRESFTVNESRTAKQYRFVLPGPSLTVAEQEQCLDELRGAAASAAFVVAS
MMAR_3482|M.marinum_M TRESFTVNESSTGEQYRFVLPGPVLAPEEQAQCLQELGFAARSAQFVVAS
TH_1728|M.thermoresistible__bu TRESLTVNEESTGHQYRFVLPGPFVTHEEQDAVLQRLREVAASAQFLVAS
MAB_1617c|M.abscessus_ATCC_199 PSGMATITVEDSGENCIVVAPGANTTFTLDE---KVHKDIICSHDVLLCQ
MAV_1743|M.avium_104 PSGTAIVVVDANAENTIVVAPGANGRFTLND---ERARAVLAGCDVMLTQ
: . . **. ::
Mflv_0751|M.gilvum_PYR-GCK GSLPPGIPASWYADITARLSGQGCRIAVDTSDAPLTALVDNLDRGAPDLI
Mvan_0095|M.vanbaalenii_PYR-1 GSLPPGVPASWYGDVVASLASRDCRVAVDTSDAPLAALIGSLDRGAPDLI
MSMEG_1378|M.smegmatis_MC2_155 GSLPAGFAPARLAAAVDEARAAGRRVAVDASGAAMTAVLAG---GSPHLI
Mb2054c|M.bovis_AF2122/97 GSLPPGVAADYYQRVADICRRSSTPLILDTSGGGLQHISSG-----VFLL
Rv2029c|M.tuberculosis_H37Rv GSLPPGVAADYYQRVADICRRSSTPLILDTSGGGLQHISSG-----VFLL
MMAR_3482|M.marinum_M GSLPPGVPADFYQRVAEACRGLGARLILDTSGGGLSHISAG-----VYLL
TH_1728|M.thermoresistible__bu GSLPPGVPADFYQRVADLCRELGVLLILDSSGGGLTQVRYG-----AFLV
MAB_1617c|M.abscessus_ATCC_199 LEIPVRVALT----AARWAAEAGKQFVLNASPVPER--------------
MAV_1743|M.avium_104 LEIPVATAVA----AARHARSGGAVVVVNASPAGRD--------------
.:* . . . . :::*
Mflv_0751|M.gilvum_PYR-GCK KPNAEELAGVLGLDADVLENAVAQGDPAPVVAAGRQMIDRG-IGAVLATL
Mvan_0095|M.vanbaalenii_PYR-1 KPNAEELAGVLGLSAQSLEDAVLQGDPGPVVAAARQLIDRG-IGAVLATL
MSMEG_1378|M.smegmatis_MC2_155 KPNLDELADATGATPATLAD---------VVDAARSLR----VDTVLVSL
Mb2054c|M.bovis_AF2122/97 KASVRELRECVGSELLTEPE---------QLAAAHELIDRGRAEVVVVSL
Rv2029c|M.tuberculosis_H37Rv KASVRELRECVGSELLTEPE---------QLAAAHELIDRGRAEVVVVSL
MMAR_3482|M.marinum_M KASVRELRECVGRPLVTEAE---------QLAAASELVETGRAEVVVVSL
TH_1728|M.thermoresistible__bu KPSKRELREYVGRDIPTTAD---------QIAAAHELIDRGVTQAVVVSL
MAB_1617c|M.abscessus_ATCC_199 SPDLAELAFRASVVVVNETE-------------ARSWLYPS--RHLVTTL
MAV_1743|M.avium_104 PDSLSELAAAADVVITNEEE-------------ASEWPWRP--RHLVVTL
. ** . : . . ::.:*
Mflv_0751|M.gilvum_PYR-GCK GAAGAVLVDKDGAWLAAPPPIVPR-STVGAGDSSLAGYLRAEVSGAAPPE
Mvan_0095|M.vanbaalenii_PYR-1 GAAGAVLVDHNGAWMATPPPIVPR-STVGAGDSSLAGYVRAEVGGAVPPQ
MSMEG_1378|M.smegmatis_MC2_155 GGDGAVLVTSDLALHGTAPVDRVV-NTVGAGDALLAGYLAACSDG--PAD
Mb2054c|M.bovis_AF2122/97 GSQGALLATRHASHRFSSIPMTAV-SGVGAGDAMVAAITVGLSRGWSLIK
Rv2029c|M.tuberculosis_H37Rv GSQGALLATRHASHRFSSIPMTAV-SGVGAGDAMVAAITVGLSRGWSLIK
MMAR_3482|M.marinum_M GSDGALLATRRARHRFRAVEMRGG-SGVGAGDAMVAAITVGLARGWPLVK
TH_1728|M.thermoresistible__bu GADGALLATADVDQKFEAIDVPVVLSGVGAGDAMVAGITVALSRRWSLSD
MAB_1617c|M.abscessus_ATCC_199 GDEGSRYRSPQGQITVPSYPVRPL-DTTGAGDVFAGVLAAWWPQGAET--
MAV_1743|M.avium_104 GSHGARYVGADGEYSVPSPDVDAV-DTTGAGDVFAGVLAANWPLEPGSPD
* *: . . .**** .
Mflv_0751|M.gilvum_PYR-GCK ----RLQMAVAYGSAAAALPGTTLPSPAEIDVDGVRVTPISPVPVTQQQL
Mvan_0095|M.vanbaalenii_PYR-1 ----RLQMAVAYGSAAAALPGTTLPSPAEIDLNAVQVSPISPIPATR---
MSMEG_1378|M.smegmatis_MC2_155 ----ALASALRFGATAVEHEGTLIGVPDPQ--RPVRIGPARGDLRLG---
Mb2054c|M.bovis_AF2122/97 ----SVRLGNAAGAAMLLTPGTAACNRDDVERFFELAAEPTEVGQDQYVW
Rv2029c|M.tuberculosis_H37Rv ----SVRLGNAAGAAMLLTPGTAACNRDDVERFFELAAEPTEVGQDQYVW
MMAR_3482|M.marinum_M ----SVRLGIAAGAAMLLTPGTQACERADVERLFEHVAEPVEVA------
TH_1728|M.thermoresistible__bu ----AVRYGVATGSAMLLTPGTAPCRPEDVERLFAQAAPPVDVTR-----
MAB_1617c|M.abscessus_ATCC_199 ----ALRRANVAGALATLRAGAGNCAPTGARIELSLKGS-----------
MAV_1743|M.avium_104 QRRLALRRACAAGALATLVPGAGDCAPRAEEIERALRGKS----------
: . *: *:
Mflv_0751|M.gilvum_PYR-GCK ----------
Mvan_0095|M.vanbaalenii_PYR-1 ----------
MSMEG_1378|M.smegmatis_MC2_155 ----------
Mb2054c|M.bovis_AF2122/97 HPIVNPEASP
Rv2029c|M.tuberculosis_H37Rv HPIVNPEASP
MMAR_3482|M.marinum_M ----------
TH_1728|M.thermoresistible__bu SPARN-----
MAB_1617c|M.abscessus_ATCC_199 ----------
MAV_1743|M.avium_104 ----------