For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSEKTRTGTDEIRYDVKGHVARVTFDRPHVLNALDAKSTQRCNEIWDQIENDPSVHVVVVTGAGEKAFCT GADMSVGGVGKTGVDYWAGLEPNGFAGLSLRETLDVPVIARVNGYALGGGMETVLGADLVIAADHATFGL TEPRVGRVPLDGGMVKLRERIPHTQAMSLLLTGRKASAREMADMGLVNEVVPMAELDAAVDRWVEQILAC APTSLRAIKQVVTRTAHLSAREAHAARLPALMEALVSPNATEGVEAFQQKRKPNWSDR
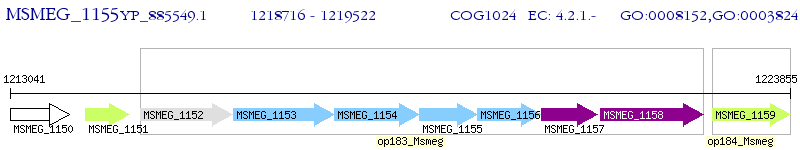
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1155 | - | - | 100% (268) | carnitinyl-CoA dehydratase |
| M. smegmatis MC2 155 | MSMEG_5198 | - | 4e-35 | 35.97% (253) | carnitinyl-CoA dehydratase |
| M. smegmatis MC2 155 | MSMEG_5915 | - | 4e-34 | 33.71% (264) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3545 | echA19 | 2e-35 | 33.20% (253) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_4354 | - | 6e-36 | 34.36% (259) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv3516 | echA19 | 5e-35 | 33.20% (253) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 5e-28 | 30.89% (259) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_0836c | - | 7e-39 | 35.14% (259) | putative enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_5002 | echA19 | 2e-38 | 36.36% (253) | enoyl-CoA hydratase EchA19 |
| M. avium 104 | MAV_2597 | - | 9e-39 | 38.43% (255) | carnitinyl-CoA dehydratase |
| M. thermoresistible (build 8) | TH_4462 | - | 3e-39 | 33.84% (263) | PUTATIVE putative enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_4076 | echA19 | 6e-39 | 36.36% (253) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_1990 | - | 2e-35 | 33.46% (257) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3545|M.bovis_AF2122/97 ---------------------MESGPDALVERRGHTLIVTMNRPAARNAL
Rv3516|M.tuberculosis_H37Rv ---------------------MESGPDALVERRGHTLIVTMNRPAARNAL
MMAR_5002|M.marinum_M ----------MAPNTSETPANGESGPDALVEQRGHTLIVTMNRPSRRNAL
MUL_4076|M.ulcerans_Agy99 MPDVMRKAVLVAPNTSETPANGESGPDALVEQRGHTLIVTMNRPSRRNAL
MAB_0836c|M.abscessus_ATCC_199 ---------------------MSAARELLVERDGPVVILTMNRPHRRNAL
Mflv_4354|M.gilvum_PYR-GCK -----------------------MSDEVLLERRDRVLIITINRPEARNAF
Mvan_1990|M.vanbaalenii_PYR-1 -----------------------MADEVLLERRDRVLIITINRPEARNAF
TH_4462|M.thermoresistible__bu --------------MTAPTTARQATDPVLVSADSHVLTITLNRPDVHNAV
MAV_2597|M.avium_104 ---------------MAETLTEVTRQAAVVERRGNVALITIDRPDARNAV
MLBr_02402|M.leprae_Br4923 ----------------------MKYDTILVDGYQRVGIITLNRPQALNAL
MSMEG_1155|M.smegmatis_MC2_155 ----------------MSEKTRTGTDEIRYDVKGHVARVTFDRPHVLNAL
. . :*::** **.
Mb3545|M.bovis_AF2122/97 STEMMRIMVQAWDRVDNDPDIRCCILTGAG-GYFCAGMDLKAATQKPPGD
Rv3516|M.tuberculosis_H37Rv STEMMRIMVQAWDRVDNDPDIRCCILTGAG-GYFCAGMDLKAATQKPPGD
MMAR_5002|M.marinum_M SGEMMQIMVEAWDRVDNDPDIRCCILTGAG-GYFCAGMDLKAATKKPPGD
MUL_4076|M.ulcerans_Agy99 SGEMMQIMVEAWDRVDNDPDIRCCILTGAG-DYFCAGMDLKAATKKPPGD
MAB_0836c|M.abscessus_ATCC_199 STNMVSQFAAAWDEIDHDDGIRAAILTGAG-SAYCVGGDLSDGWMVRDGS
Mflv_4354|M.gilvum_PYR-GCK NLAVAQGLADAMDELDDTPELSVAIITGAG-GNFCAGMDLKAFMA---GE
Mvan_1990|M.vanbaalenii_PYR-1 NLAVAQGLADAMDELDSSAELSVAIITGAG-GNFCAGMDLKAFMA---GE
TH_4462|M.thermoresistible__bu NLDMAQRISDALDRLDDDPDLRVGIIAANG-RSFCAGMDLKAFAR---GE
MAV_2597|M.avium_104 NGAVSTAVGDALEEAQRDPEVWAVVITGAGDKSFCAGADLKAISR---GE
MLBr_02402|M.leprae_Br4923 NSQMMNEITNAAKELDIDPDVGAILITGSP-KVFAAGADIKEMASLTFTD
MSMEG_1155|M.smegmatis_MC2_155 DAKSTQRCNEIWDQIENDPSVHVVVVTGAGEKAFCTGADMSVGGVGKTGV
. .. : : :::. :..* *:.
Mb3545|M.bovis_AF2122/97 SFKDGSYDPSRIDA-LLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRVA
Rv3516|M.tuberculosis_H37Rv SFKDGSYGPSRIDA-LLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRVA
MMAR_5002|M.marinum_M SFKDGSYDPSRIDA-LLKGRRLKKPLIAAVEGPAIAGGTEILQGTDIRVA
MUL_4076|M.ulcerans_Agy99 SFKDGSYDPSRIDA-LLKGRRLKKPLIAAVEGPAIAGGTEILQGTDIRVA
MAB_0836c|M.abscessus_ATCC_199 AP---PLDPATIGKGLLLSHTLTKPLIAAVNGACLGGGCEMLQQTDIRVS
Mflv_4354|M.gilvum_PYR-GCK VPAIEGR---GIG---FTERPPRKPLIAAVEGYALAGGTEIVLATDLIVA
Mvan_1990|M.vanbaalenii_PYR-1 VPAIEGR---GIG---FTERPPRKPVIAAVEGYALAGGTEIVLATDLIVA
TH_4462|M.thermoresistible__bu VPRPEPR---GFAG--IVKQPPAKPLVAAVEGAAVGGGFEIVLACDLIVA
MAV_2597|M.avium_104 NLYHAEHPEWGFAG--YVHHFIDKPTIAAVNGTALGGGSELALASDLVIA
MLBr_02402|M.leprae_Br4923 AFDADFFSAWGKLA------AVRTPMIAAVAGYALGGGCELAMMCDLLIA
MSMEG_1155|M.smegmatis_MC2_155 DYWAGLE-PNGFAG-LSLRETLDVPVIARVNGYALGGGMETVLGADLVIA
* :* * * .:.** * *: ::
Mb3545|M.bovis_AF2122/97 GESAKFGISEAKWSLYPMGGSAVRLVRQIPYTLACDLLLTGRHITAAEAK
Rv3516|M.tuberculosis_H37Rv GESAKFGISEAKWSLYPMGGSAVRLVRQIPYTLACDLLLTGRHITAAEAK
MMAR_5002|M.marinum_M AESAKFGISEAKWSLYPMGGSAVRLVRQIPYTVACDLLLTGRHITAAEAK
MUL_4076|M.ulcerans_Agy99 AESAKFGISEAEWSLYPMGGSAVRLVRQIPYTVACDLLLTGRHITAAEAK
MAB_0836c|M.abscessus_ATCC_199 DEHATFGLPEVQRGLVPGAGSMVRLKRQIPYTKAMEMILTGEPLTAFEAY
Mflv_4354|M.gilvum_PYR-GCK AKNAKFGIPEVKRGLVAAGGGLLRLQKRIPYQKALELALTGDSFTAEEAS
Mvan_1990|M.vanbaalenii_PYR-1 AKNAKFGIPEVKRGLVAAGGGLLRLQKRIPYQKALELALTGDSFTAEEAS
TH_4462|M.thermoresistible__bu SESARFSLPEVTRGLTPNGGGLLRLPRRIPHHLAMELILTGRPVTAARAA
MAV_2597|M.avium_104 CESASFGLPEVKRGLIAGAGGVFRIVEQLPRKVALELVLTGEPMTASDAL
MLBr_02402|M.leprae_Br4923 ADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAE
MSMEG_1155|M.smegmatis_MC2_155 ADHATFGLTEPRVGRVPLDGGMVKLRERIPHTQAMSLLLTGRKASAREMA
. * *. .* . . *. :: . : * .: *** *
Mb3545|M.bovis_AF2122/97 EMGLIGHVVPDGQALTKALELADAISANGPLAVQAILRSIRETEC---MP
Rv3516|M.tuberculosis_H37Rv EMGLIGHVVPDGQALTKALELADAISANGPLAVQAILRSIRETEC---MP
MMAR_5002|M.marinum_M EMGLVGHVVPDGQALTKALEIAEIIAANGPLAVQAILRTIRETEG---MH
MUL_4076|M.ulcerans_Agy99 EMGLVGHVVPDGQALTKALEIAEIIAANGPLAVQAILRTIRETEG---MH
MAB_0836c|M.abscessus_ATCC_199 HFGLVGHVVPAGTALDKARSLADRIVRNGPLAVRNAKEAIVRSGW---LA
Mflv_4354|M.gilvum_PYR-GCK AWGFVNVLTEPGGALDGALALAERITANGPLAVAATKEVIAKSAD---WT
Mvan_1990|M.vanbaalenii_PYR-1 QWGFVNVLTEPGEALNGALALAERITANGPLAVAVTKEVIVKSAD---WT
TH_4462|M.thermoresistible__bu ELGLLNRLTPPGSALSTARELAAVIAANGPLAIRAAKRVVEESPD---WP
MAV_2597|M.avium_104 RWGLINEVVPDGTVVEAALALAERITCNAPLSVQASKRVAYGADDGIIGA
MLBr_02402|M.leprae_Br4923 RSGLVSRVVLADDLLPEAKAVATTISQMSRSATRMAKEAVNRSFES---T
MSMEG_1155|M.smegmatis_MC2_155 DMGLVNEVVPMAELDAAVDRWVEQILACAPTSLRAIKQVVTRTAH---LS
*::. :. . . * . : . :
Mb3545|M.bovis_AF2122/97 ENEAFKIDTQIGIKVFLSDDAKEGPRAFAEKRAPNFQNR
Rv3516|M.tuberculosis_H37Rv ENEAFKIDTQIGIKVFLSDDAKEGPRAFAEKRAPNFQNR
MMAR_5002|M.marinum_M ENEAFKIDTRIGIEVFLSDDAKEGPQAFAQKRKPNFQNR
MUL_4076|M.ulcerans_Agy99 ENEAFKIDTRIGIEVFLSDDAKEGPQAFAQKRKPNFQNR
MAB_0836c|M.abscessus_ATCC_199 EEDARAIEARLTRPVITSADAREGLAAFKEKREARFTGR
Mflv_4354|M.gilvum_PYR-GCK EAEMWKKQGEIVFPVFSSKDAMEGATAFAEKRKPNWTGT
Mvan_1990|M.vanbaalenii_PYR-1 EAEMWKKQGELVMSVFSSKDAMEGATAFAEKRKPNWTGS
TH_4462|M.thermoresistible__bu RTEMFTRQEDIVAPVRSSADAAEGARAFAEKRMPLWRNR
MAV_2597|M.avium_104 EEPKWERTIREFTELLKSEDAKEGPLAFAEKRQPVWKAR
MLBr_02402|M.leprae_Br4923 LAEGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR
MSMEG_1155|M.smegmatis_MC2_155 AREAHAARLPALMEALVSPNATEGVEAFQQKRKPNWSDR
: : ** ** :** . :