For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAPNTSETPANGESGPDALVEQRGHTLIVTMNRPSRRNALSGEMMQIMVEAWDRVDNDPDIRCCILTGAG GYFCAGMDLKAATKKPPGDSFKDGSYDPSRIDALLKGRRLKKPLIAAVEGPAIAGGTEILQGTDIRVAAE SAKFGISEAKWSLYPMGGSAVRLVRQIPYTVACDLLLTGRHITAAEAKEMGLVGHVVPDGQALTKALEIA EIIAANGPLAVQAILRTIRETEGMHENEAFKIDTRIGIEVFLSDDAKEGPQAFAQKRKPNFQNR
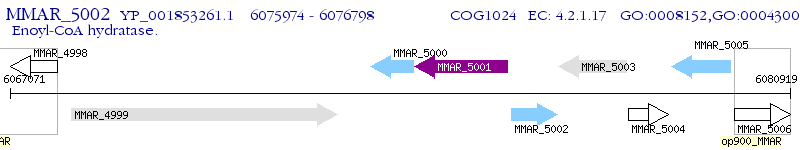
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5002 | echA19 | - | 100% (274) | enoyl-CoA hydratase EchA19 |
| M. marinum M | MMAR_0465 | echA1 | 4e-52 | 43.66% (268) | enoyl-CoA hydratase EchA1 |
| M. marinum M | MMAR_4317 | echA1_1 | 2e-45 | 38.89% (270) | enoyl-CoA hydratase, EchA1_1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3545 | echA19 | 1e-137 | 91.22% (262) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_1542 | - | 1e-126 | 85.33% (259) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv3516 | echA19 | 1e-137 | 90.84% (262) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 3e-34 | 37.01% (254) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_4164 | - | 1e-118 | 78.38% (259) | enoyl-CoA hydratase |
| M. avium 104 | MAV_0643 | - | 1e-139 | 89.96% (269) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5915 | - | 1e-127 | 86.05% (258) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_1820 | echA19 | 1e-124 | 84.17% (259) | POSSIBLE ENOYL-CoA HYDRATASE ECHA19 (ENOYL HYDRASE) |
| M. ulcerans Agy99 | MUL_4076 | echA19 | 1e-155 | 99.27% (274) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_5216 | - | 1e-125 | 84.23% (260) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5002|M.marinum_M -------------MAPNTSETPANGESGPDALVEQRGHTLIVTMNRPSRR
MUL_4076|M.ulcerans_Agy99 ---MPDVMRKAVLVAPNTSETPANGESGPDALVEQRGHTLIVTMNRPSRR
Mb3545|M.bovis_AF2122/97 ------------------------MESGPDALVERRGHTLIVTMNRPAAR
Rv3516|M.tuberculosis_H37Rv ------------------------MESGPDALVERRGHTLIVTMNRPAAR
MAV_0643|M.avium_104 -----------------MAEASANEKTGPDALVEQRGHTLIVTLNRPHAR
MSMEG_5915|M.smegmatis_MC2_155 ------------------MSE---TGKGPDALVEQRGHTLIVTLNRPEAR
Mvan_5216|M.vanbaalenii_PYR-1 ------------------MSD---PQKGPDALVEQRGHTLIVTLNRPEAR
Mflv_1542|M.gilvum_PYR-GCK MLHFQTSLWHRRWKADQFVSDEQNPEKAPDALVEQRGHTLIVTMNRPEKR
TH_1820|M.thermoresistible__bu --------------VDQPVSA---LGSQPHALIEQRGHTLILTLNRPEAR
MAB_4164|M.abscessus_ATCC_1997 -------MTEADALRASGSSHTVTEQDAPHALVELRDHVLIVTMNRPHAR
MLBr_02402|M.leprae_Br4923 -------------------------MKYDTILVDGYQRVGIITLNRPQAL
*:: :. *:*:***
MMAR_5002|M.marinum_M NALSGEMMQIMVEAWDRVDNDPDIRCCILTGAGGYFCAGMDLKAATKKPP
MUL_4076|M.ulcerans_Agy99 NALSGEMMQIMVEAWDRVDNDPDIRCCILTGAGDYFCAGMDLKAATKKPP
Mb3545|M.bovis_AF2122/97 NALSTEMMRIMVQAWDRVDNDPDIRCCILTGAGGYFCAGMDLKAATQKPP
Rv3516|M.tuberculosis_H37Rv NALSTEMMRIMVQAWDRVDNDPDIRCCILTGAGGYFCAGMDLKAATQKPP
MAV_0643|M.avium_104 NALSTEMMEIMVQAWDRVDNDDDIRCCILTGAGGYFCAGMDLKAATKKPP
MSMEG_5915|M.smegmatis_MC2_155 NALSGEMLSIMVEAWDRVDNDPEIRTCILTGAGGYFCSGMDLKGADKKPP
Mvan_5216|M.vanbaalenii_PYR-1 NALSTEMLSIMVDAWNRVDEDPEIRTCILTGAGGYFCAGMDLKAASKKPP
Mflv_1542|M.gilvum_PYR-GCK NALSAEMMQIMVEAWDRVDSDPEIRSCILTGAGGAFCAGMDLKKADSQAP
TH_1820|M.thermoresistible__bu NALSTEMLATMVEAWDRVDNDPEIRTCILTGAGGYFCSGMDLKAATARPP
MAB_4164|M.abscessus_ATCC_1997 NALSGEMLEIMTQAWDRVDNDPEVRVCILTGAGGYFCAGADLKAMNKRAP
MLBr_02402|M.leprae_Br4923 NALNSQMMNEITNAAKELDIDPDVGAILITGSPKVFAAGADIKEMASLTF
***. :*: :.:* ..:* * :: ::**: *.:* *:* .
MMAR_5002|M.marinum_M GDSFKDGSYDPSRIDALLKGRRLKKPLIAAVEGPAIAGGTEILQGTDIRV
MUL_4076|M.ulcerans_Agy99 GDSFKDGSYDPSRIDALLKGRRLKKPLIAAVEGPAIAGGTEILQGTDIRV
Mb3545|M.bovis_AF2122/97 GDSFKDGSYDPSRIDALLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRV
Rv3516|M.tuberculosis_H37Rv GDSFKDGSYGPSRIDALLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRV
MAV_0643|M.avium_104 GESFKDGSYDPSRIDALLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRV
MSMEG_5915|M.smegmatis_MC2_155 GDSFKDGSYDPSKIPGLLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRV
Mvan_5216|M.vanbaalenii_PYR-1 GDSFADGSYDPSKIEGLLKGRRLKKPLIAAVEGPAIAGGTEILQGTDIRV
Mflv_1542|M.gilvum_PYR-GCK GDSFKDGSYDPSKIPALLKGRRLKKPLIAAVEGAAIAGGTEILQGTDIRV
TH_1820|M.thermoresistible__bu GDSFKDGSYDPSTIPALLKGRRLTKPLIAAVEGPAIAGGTEILQGTDIRI
MAB_4164|M.abscessus_ATCC_1997 GDQFSDGSYDPSVIPGLLKGRRLTKPLIAAVEGPAIAGGTEILQATDIRV
MLBr_02402|M.leprae_Br4923 TDAFDADFFS-----AWGKLAAVRTPMIAAVAGYALGGGCELAMMCDLLI
: * . :. . * : .*:**** * *:.** *: *: :
MMAR_5002|M.marinum_M AAESAKFGISEAKWSLYPMGGSAVRLVRQIPYTVACDLLLTGRHITAAEA
MUL_4076|M.ulcerans_Agy99 AAESAKFGISEAEWSLYPMGGSAVRLVRQIPYTVACDLLLTGRHITAAEA
Mb3545|M.bovis_AF2122/97 AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTLACDLLLTGRHITAAEA
Rv3516|M.tuberculosis_H37Rv AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTLACDLLLTGRHITAAEA
MAV_0643|M.avium_104 AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTVACDLLLTGRHITAAEA
MSMEG_5915|M.smegmatis_MC2_155 AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTIACDMLLTGRHITAAEA
Mvan_5216|M.vanbaalenii_PYR-1 AGESAKFGISEAKWSLYPMGGSAVRLPRQIPYTIACDLLLTGRHITAAEA
Mflv_1542|M.gilvum_PYR-GCK AGESAKFGVSEAKWSLYPMGGSAVRLVRQIPYTVACDILLTGRHISAAEA
TH_1820|M.thermoresistible__bu AGESAKFGISEAKWSLYPMGGSAVRLVRQIPYTIAADLLLTGRHITAAEA
MAB_4164|M.abscessus_ATCC_1997 AGESAKFGVSEVKWSLYPMGGSAVRLPRQIPYTVACDILLTGRHIKAPEA
MLBr_02402|M.leprae_Br4923 AADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEA
*.::**** .* : .: * *.: ** * * : * *::**** * *.**
MMAR_5002|M.marinum_M KEMGLVGHVVPDGQALTKALEIAEIIAANGPLAVQAILRTIRETEGMHEN
MUL_4076|M.ulcerans_Agy99 KEMGLVGHVVPDGQALTKALEIAEIIAANGPLAVQAILRTIRETEGMHEN
Mb3545|M.bovis_AF2122/97 KEMGLIGHVVPDGQALTKALELADAISANGPLAVQAILRSIRETECMPEN
Rv3516|M.tuberculosis_H37Rv KEMGLIGHVVPDGQALTKALELADAISANGPLAVQAILRSIRETECMPEN
MAV_0643|M.avium_104 KEMGLIGHVVPDGQALAKALEIAEVIENNGPLAVQAILRAIRETEGMHEN
MSMEG_5915|M.smegmatis_MC2_155 KEYGLIGYVVPDGTALDKALEIAETINNNGPLAVQAILKTIRETEGMHED
Mvan_5216|M.vanbaalenii_PYR-1 LQYGLIGHVVPDGTALDKALEIAEVINNNGPLAVQAILKTIRETEGMHEE
Mflv_1542|M.gilvum_PYR-GCK KEFGLIGHVVPDGQALTKALEIAEVINGNGPLAVQAILKTIRETEGMHEE
TH_1820|M.thermoresistible__bu KEIGLIGYVVPDGTALDKALEIAEVINNNGPLAVQAILRSIRETEGLPEN
MAB_4164|M.abscessus_ATCC_1997 KDIGLIGHVVPDGQALEKALELANMIAGNGPLAVQAVLKTIRDSEGMHEN
MLBr_02402|M.leprae_Br4923 ERSGLVSRVVLADDLLPEAKAVATTISQMSRSATRMAKEAVNRSFESTLA
**:. ** . * :* :* * . *.: .::. :
MMAR_5002|M.marinum_M EAFKIDTRIGIEVFLSDDAKEGPQAFAQKRKPNFQNR
MUL_4076|M.ulcerans_Agy99 EAFKIDTRIGIEVFLSDDAKEGPQAFAQKRKPNFQNR
Mb3545|M.bovis_AF2122/97 EAFKIDTQIGIKVFLSDDAKEGPRAFAEKRAPNFQNR
Rv3516|M.tuberculosis_H37Rv EAFKIDTQIGIKVFLSDDAKEGPRAFAEKRAPNFQNR
MAV_0643|M.avium_104 EAFKIDTQIGIKVFLSEDAKEGPRAFAEKRKPNFQNR
MSMEG_5915|M.smegmatis_MC2_155 EAFGPDTRNGIPVFLSEDAKEGPRAFKEKRAPNFQMK
Mvan_5216|M.vanbaalenii_PYR-1 EAFKPDTANGIPVFLSEDAKEGPRAFKEKRAPNFQMK
Mflv_1542|M.gilvum_PYR-GCK EAFKPDTANGIPVFLSEDSKEGPKAFLEKRKPVWKLK
TH_1820|M.thermoresistible__bu EAFKIDTKIGIEVFLSEDAKEGPRAFKEKRKPVFKMK
MAB_4164|M.abscessus_ATCC_1997 EAFKADTKVGIGVFTSNDAKEGPRAFAEKRAPNFTGS
MLBr_02402|M.leprae_Br4923 EGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR
*.: : .* ::* .** ** :** * :