For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDTPNPDAGRAVADTGLLVAAIRAAESRREDRLFEDPFAEKLAGETGRRMLEEALATTGDKSTLQIVVRT KFWDEALLRATTHVDQVVLLAAGMDARAFRLAWPAGTVVYELDQPEVIAAKSGLLADASPRCRWTALGVD LTRDWTTALRSTTFDPARPAVWLAEGLLQYLDETAVHTLFDRVDAMSAPGSVLLYDVVGKTLLDAAMLAA VRESMARSGAPWRFGTDAPGELSTRLGWSANVTDIAEPGNAYGRWFTPAVPLDVPGVPRGYFVEATKPG*
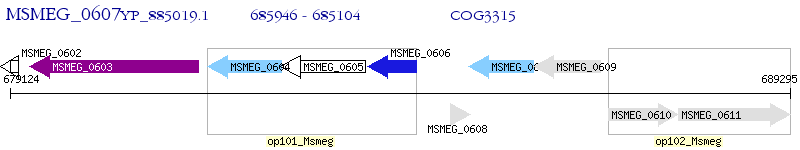
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0607 | - | - | 100% (280) | methyltransferase, putative, family protein |
| M. smegmatis MC2 155 | MSMEG_1480 | - | 2e-37 | 38.61% (259) | methyltransferase |
| M. smegmatis MC2 155 | MSMEG_1481 | - | 7e-37 | 36.52% (282) | methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0917c | - | 4e-38 | 38.51% (296) | hypothetical protein Mb0917c |
| M. gilvum PYR-GCK | Mflv_5025 | - | 1e-38 | 40.86% (301) | putative methyltransferase |
| M. tuberculosis H37Rv | Rv0893c | - | 4e-38 | 38.51% (296) | hypothetical protein Rv0893c |
| M. leprae Br4923 | MLBr_02640 | - | 3e-34 | 40.69% (204) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_4607c | - | 3e-38 | 37.32% (276) | hypothetical protein MAB_4607c |
| M. marinum M | MMAR_0955 | - | 2e-37 | 35.76% (302) | hypothetical protein MMAR_0955 |
| M. avium 104 | MAV_0219 | - | 1e-105 | 69.29% (267) | methyltransferase, putative, family protein |
| M. thermoresistible (build 8) | TH_4502 | - | 1e-36 | 41.67% (216) | PUTATIVE methyltransferase |
| M. ulcerans Agy99 | MUL_0706 | - | 1e-36 | 35.43% (302) | hypothetical protein MUL_0706 |
| M. vanbaalenii PYR-1 | Mvan_1344 | - | 9e-37 | 45.32% (203) | putative methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0955|M.marinum_M --MARTEDDSWEITESVGATALGVASARAAETRSQHPLISDPFAQVFLDA
MUL_0706|M.ulcerans_Agy99 --MARTEDDSWEITESVGATALGVASARAAETRSQHPLISDPFAQVFLDA
Mb0917c|M.bovis_AF2122/97 ---MRTEDDSWDVTTSVGSTGLLVAAARALETQKADPLAIDPYAEVFCRA
Rv0893c|M.tuberculosis_H37Rv ---MRTEDDSWDVTTSVGSTGLLVAAARALETQKADPLAIDPYAEVFCRA
Mflv_5025|M.gilvum_PYR-GCK --MARTPDDSWDLASSVGATATMVAAGRAVASADPDPLINDPYAEPLVRA
Mvan_1344|M.vanbaalenii_PYR-1 --MARTADDSWDPASSVGATATMVAAGRAAASADPDPLINDPYAEPLVRA
MLBr_02640|M.leprae_Br4923 ---MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLLVTN
TH_4502|M.thermoresistible__bu MSSLRTDDDTWDIATSVGSTAVMVAASRAAETEREDALIRDPYAKVLVAG
MAB_4607c|M.abscessus_ATCC_199 ---MRVDGDTWDITSSVGATALGVAAARATETLRADALIRDPFAQILVDA
MSMEG_0607|M.smegmatis_MC2_155 ----MGDTPNPDAGRAVADTGLLVAAIRAAESRREDRLFEDPFAEKLAGE
MAV_0219|M.avium_104 ----MMDAN----LAAVADTALLVAAIRAHETTRDDRLFADPFAARLAGD
:*. *. **: ** : . * **:* :
MMAR_0955|M.marinum_M VGDG-VWNWHSAPQLPAELLEIEPDLPLQMEAMVSYMASRTAFFDEFFLD
MUL_0706|M.ulcerans_Agy99 VGDG-VWNWHSAPQLPAELLEIEPDLPLQMEAMVSYMASRTAFFDEFFLD
Mb0917c|M.bovis_AF2122/97 AG-G-EWADVLDGKLPDHYLTTG-DFG---EHFVNFQGARTRYFDEYFSR
Rv0893c|M.tuberculosis_H37Rv AG-G-EWADVLDGKLPDHYLTTG-DFG---EHFVNFQGARTRYFDEYFSR
Mflv_5025|M.gilvum_PYR-GCK VGLD-FFTRMLDG--ELDLSVFPDSSPERAQAMIDGMAVRTRFFDDCCLA
Mvan_1344|M.vanbaalenii_PYR-1 VGLS-FFTKMLDG--ELDLSQFADGSPERVQAMIDGMAVRTRFFDDCCGT
MLBr_02640|M.leprae_Br4923 TGAGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTYFNN
TH_4502|M.thermoresistible__bu AGTG-VWELILDDDFTAKIEAADPETAAIFDHMGNYQAVRTHFFDTYFAR
MAB_4607c|M.abscessus_ATCC_199 TGKATGWERLVAG----DIDWPDPEAGRIYDRMVDYQATRTHFFDEYFLA
MSMEG_0607|M.smegmatis_MC2_155 TGRR----------------MLEEALATTGDKSTLQIVVRTKFWDEALLR
MAV_0219|M.avium_104 RGHE----------------LLAGALAATGESATAQIVVRTRFWDDALLE
* : ** ::*
MMAR_0955|M.marinum_M ATRAGIGQAVILAAGLDSRAWRLPWPAGTTVFELDQPRVLEFKAATLAEH
MUL_0706|M.ulcerans_Agy99 ATRAGIGQAVILAAGLDSRAWRLPWPAGTTVFELDQPRVLEFKAATLAEH
Mb0917c|M.bovis_AF2122/97 ATAAGMKQVVILAAGLDSRAFRLQWPIGTTIFELDRPQVLDFKNAVLADY
Rv0893c|M.tuberculosis_H37Rv ATAAGMKQVVILAAGLDSRAFRLQWPIGTTIFELDRPQVLDFKNAVLADY
Mflv_5025|M.gilvum_PYR-GCK AASAGVRQVVILAAGLDARTYRLPWPDGTVVYELDQPDVIAFKTQTLRNL
Mvan_1344|M.vanbaalenii_PYR-1 STTAGITQVVILASGLDARAYRLGWPAGTVVYELDQPAVIEFKTRTLAGL
MLBr_02640|M.leprae_Br4923 AVIDGIRQFVILASGLDSRAYRLDWPTGTTVYEIDQPKVLAYKSTTLAEH
TH_4502|M.thermoresistible__bu AAEAGIRQVVILASGLDSRAYRLPWPDGTVVYEIDQPKVLEYKSATLAAH
MAB_4607c|M.abscessus_ATCC_199 AAAAGIRQVVILASGLDSRAYRLDWPAGTTVYEIDQPQVLKFKDSALAAH
MSMEG_0607|M.smegmatis_MC2_155 ATTH-VDQVVLLAAGMDARAFRLAWPAGTVVYELDQPEVIAAKSGLLADA
MAV_0219|M.avium_104 AAQQ-ISQVVILAAGMDARAYRLAWPDGTVVYELDQPHVLSAKDGVLAGE
:. : * *:**:*:*:*::** ** **.::*:*:* *: * *
MMAR_0955|M.marinum_M GAEPACGRVAVAVDLRQDWPTALRQAGFDPSVPSVWSAEGLMPYLPAVAQ
MUL_0706|M.ulcerans_Agy99 GAEPACGRVAVAVDLRQDWPTALRQAGFDPSVSSVWSAEGLMPYLPAVAQ
Mb0917c|M.bovis_AF2122/97 HIRPRAQRRSVAVDLRDEWQIALCNNGFDANRPSAWIAEGLLVYLSAEAQ
Rv0893c|M.tuberculosis_H37Rv HIRPRAQRRSVAVDLRDEWQIALCNNGFDANRPSAWIAEGLLVYLSAEAQ
Mflv_5025|M.gilvum_PYR-GCK GAEPAATQRPVPVDLREDWPAALRAAGFDATRPTAWLAEGLLIYLPPEAQ
Mvan_1344|M.vanbaalenii_PYR-1 GAHPTATHRPVPIDLREDWPAALHAAGFDASRPTAWLAEGLLIYLPPEAQ
MLBr_02640|M.leprae_Br4923 GVTPTADRREVPIDLRQDWPPALRSAGFDPSARTAWLAEGLLMYLPATAQ
TH_4502|M.thermoresistible__bu GVQPAADRREVAVDLRHDWPKALRAKGFDPQQRTAWLAEGLLMYLPADAQ
MAB_4607c|M.abscessus_ATCC_199 --QPTAQRRGVAIDLREDWPAALRAAGFDSAQPTAWLAEGLLPYLPADAQ
MSMEG_0607|M.smegmatis_MC2_155 --SPRCRWTALGVDLTRDWTTALRSTTFDPARPAVWLAEGLLQYLDETAV
MAV_0219|M.avium_104 --RPACRRVAVGVDLAQDWPAALRRAGLDPSAPAVWLIEGLLQYLDEAAV
* . : :** :* ** :*. :.* ***: ** *
MMAR_0955|M.marinum_M DLLFERVQGLTVRASRIAVEALGPKFLDPQVRANRSARMERIRAVMAHVE
MUL_0706|M.ulcerans_Agy99 DLLFERVQGLTVRASRIAVEALGPKFLDPQVRANRSARIERIRAVMAHVE
Mb0917c|M.bovis_AF2122/97 QRLFIGIDTLASPGSHVAVEEATP--LDP---CEFAAKLERERAANAQGD
Rv0893c|M.tuberculosis_H37Rv QRLFIGIDTLASPGSHVAVEEATP--LDP---CEFAAKLERERAANAQGD
Mflv_5025|M.gilvum_PYR-GCK DALFDTVTALSAPGSTLATEYVP-----GIVDFDGAKARALSEPLREHG-
Mvan_1344|M.vanbaalenii_PYR-1 DQLFDRITALSAPGSTIATEYAP-----GIIDFDDEKARAMSAPMREMG-
MLBr_02640|M.leprae_Br4923 DGLFTEIGGLSAVGSRIAVETSP----LHGDEWREQMQLRFRRVSDALG-
TH_4502|M.thermoresistible__bu DRLFENITALSAPGSRLAVETAA----EEPVDRREQMRARMERIASQFG-
MAB_4607c|M.abscessus_ATCC_199 DNLFRDIGVLSAAGSRVAVEGYEG---KLVLDESEEEAVRREQVRAAFKT
MSMEG_0607|M.smegmatis_MC2_155 HTLFDRVDAMSAPGSVLLYDVVG------KTLLDAAMLAAVRESMARSG-
MAV_0219|M.avium_104 TALLDRVDALSARASVLLYDVVG------KALLESDFMAPVLESMARSG-
*: : :: .* : :
MMAR_0955|M.marinum_M PQREIPRTDELWYFE-EREDVGDWFRRHDWDVTVTPSGELMAGYGRA---
MUL_0706|M.ulcerans_Agy99 PQREIPRTDELWYFE-EREDVGDWFRRHDWDVTVTPSGELMAGYGRA---
Mb0917c|M.bovis_AF2122/97 PR----RFFQMVYNE-RWARATEWFDERGWRATATPLAEYLRRVGRA---
Rv0893c|M.tuberculosis_H37Rv PR----RFFQMVYNE-RWARATEWFDERGWRATATPLAEYLRRVGRA---
Mflv_5025|M.gilvum_PYR-GCK ---LDIDMPSLVYTG-ARTPVMDHLRGAGWQVSGTARAELFARFGRP---
Mvan_1344|M.vanbaalenii_PYR-1 ---LDIDMPSLVYAG-TRSHVMDYLAGQGWEVTGVPRRELFTRYGRP---
MLBr_02640|M.leprae_Br4923 -FEQAVDVQELIYHDENRAVVADWLNRHGWRATAQSAPDEMRRVGRW---
TH_4502|M.thermoresistible__bu -LEKALDVAQLMYDDPDRADVPEWLTGHGWRTEWVSAPDEMRRLDRFVAA
MAB_4607c|M.abscessus_ATCC_199 AIDVDVTIENLIYEDENRADPAEWLDAHGWSVTKTDAHDEMARLGRP---
MSMEG_0607|M.smegmatis_MC2_155 --------APWRFGT---DAPGELSTRLGWSANVTDIAEPGNAYGRWF--
MAV_0219|M.avium_104 --------APWRFGT---DDPGGLCERLGWSATVTDVAEPGNRYQRWY--
: .* . : *
MMAR_0955|M.marinum_M ----AAAQVEDRVPTN---LFVAAQRK-----------------------
MUL_0706|M.ulcerans_Agy99 ----AAAQVEDRVPTN---LFVAAQRN-----------------------
Mb0917c|M.bovis_AF2122/97 ----VPEADTEAAPMVTAITFVSAVRTGLVADPARTSPSSTSIGFKRFEA
Rv0893c|M.tuberculosis_H37Rv ----VPEADTEAAPMVTAITFVSAVRTGLVADPARTSPSSTSIGFKRFEA
Mflv_5025|M.gilvum_PYR-GCK -----LPQDADGTDPLGEIVYVSATLAADA--------------------
Mvan_1344|M.vanbaalenii_PYR-1 -----LPPEAGDTDPLGEIVYVSAHRPQ----------------------
MLBr_02640|M.leprae_Br4923 ----GDGVPMAD-DKDAFAEFVTAHRL-----------------------
TH_4502|M.thermoresistible__bu AMPPGDDADAADLDDTAFSKFVTAERPS----------------------
MAB_4607c|M.abscessus_ATCC_199 --------VAEDVERGTFRGQLIQGELR----------------------
MSMEG_0607|M.smegmatis_MC2_155 -----TPAVPLDVPGVPRGYFVEATKPG----------------------
MAV_0219|M.avium_104 -----APAVPMDVAGVPRGYFVQATKQAAGD-------------------
:
MMAR_0955|M.marinum_M -
MUL_0706|M.ulcerans_Agy99 -
Mb0917c|M.bovis_AF2122/97 D
Rv0893c|M.tuberculosis_H37Rv D
Mflv_5025|M.gilvum_PYR-GCK -
Mvan_1344|M.vanbaalenii_PYR-1 -
MLBr_02640|M.leprae_Br4923 -
TH_4502|M.thermoresistible__bu -
MAB_4607c|M.abscessus_ATCC_199 -
MSMEG_0607|M.smegmatis_MC2_155 -
MAV_0219|M.avium_104 -