For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTDNFERAGAPGELRSNFTPGSTRWAVRRAQWSAMGIDWRDQEKPKIAIVNTSSKLSVCFAHLDDVAVRV AEAVRAAGGLPFEIRTTAPSDFVTSAGRKARYLMPTRDLIVNDVEAAVEGAVLDGVVFLSSCDKTTPAHL MAAARLDLPAIVVIGGYQRGGTWSGCSVDIDTVYESVGALTAGTMTVDQLGELADQAIEGPGVCAGLATA NTMHCLAEALGMTVAGSAPVRANSDRMLDNAAAAGHHIVKMVEQNLTARQVITEKSIENAIEVALALGGS VNCIRHLAAICAEADLDLDVVGMFEKHGGDAVQLAAIRPNGAHQVWDLDGVGGIQTVMRTLLPRLHGDAL TVDGRTVSERAADAQPADGDVLHPLDDPINDEPGLIILRGSLAPDGSIVKVAGVGKATRRQFNGPAKVFV GEDDAIAALGDGRIQRGDVIVLRGMGPRGGPGTVFAASFVAALNGAGLGGHVAVVTDGELSGLNHGLVVG QVMPEAADGGPLAGLQNGDTVAIDLDARRIDSHPIRDGEPVRATGEHPERGWLGQYAALVGPVQRGAVLR RPRN
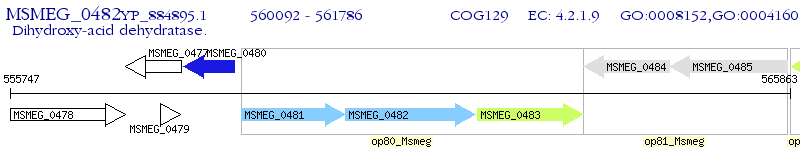
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0482 | - | - | 100% (564) | dihydroxy-acid dehydratase |
| M. smegmatis MC2 155 | MSMEG_0229 | ilvD | 5e-93 | 37.06% (572) | dihydroxy-acid dehydratase |
| M. smegmatis MC2 155 | MSMEG_1290 | ilvD | 2e-73 | 32.29% (607) | dihydroxy-acid dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0195c | ilvD | 3e-97 | 38.46% (572) | dihydroxy-acid dehydratase |
| M. gilvum PYR-GCK | Mflv_0486 | - | 4e-94 | 37.94% (572) | dihydroxy-acid dehydratase |
| M. tuberculosis H37Rv | Rv0189c | ilvD | 3e-97 | 38.46% (572) | dihydroxy-acid dehydratase |
| M. leprae Br4923 | MLBr_02608 | ilvD | 2e-94 | 38.11% (530) | dihydroxy-acid dehydratase |
| M. abscessus ATCC 19977 | MAB_4545 | - | 1e-94 | 37.41% (572) | dihydroxy-acid dehydratase |
| M. marinum M | MMAR_0432 | ilvD | 1e-95 | 37.94% (572) | dihydroxy-acid dehydratase IlvD |
| M. avium 104 | MAV_4989 | ilvD | 2e-96 | 38.29% (572) | dihydroxy-acid dehydratase |
| M. thermoresistible (build 8) | TH_0983 | ilvD | 1e-98 | 38.38% (581) | PROBABLE DIHYDROXY-ACID DEHYDRATASE ILVD (DAD) |
| M. ulcerans Agy99 | MUL_1082 | ilvD | 1e-95 | 38.11% (572) | dihydroxy-acid dehydratase |
| M. vanbaalenii PYR-1 | Mvan_0173 | - | 9e-96 | 38.11% (572) | dihydroxy-acid dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0432|M.marinum_M --------------------------MPEADIKPRSRDVTDGLEKAAARG
MUL_1082|M.ulcerans_Agy99 --------------------------MPEADIKPRSRDVTDGLEKAAARG
Mb0195c|M.bovis_AF2122/97 ---------------MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARG
Rv0189c|M.tuberculosis_H37Rv ---------------MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARG
MLBr_02608|M.leprae_Br4923 --------------------------MMPPDIKPRSRDVTDGLEKAAARG
MAV_4989|M.avium_104 ---------------MPTTDSARAADIKQPDIKPRSRDVTDGLEKAAARG
TH_0983|M.thermoresistible__bu ------------------------MAQNQPDIKPRSRDVTDGLEKAAARG
Mvan_0173|M.vanbaalenii_PYR-1 -------------------------MPSSPDIKPRSRDVTDGLEKAAARG
Mflv_0486|M.gilvum_PYR-GCK MWPMNRRAHNHPMPSDSRSHSLRASGSS-VDIKPRSRDVTDGLERTAARG
MAB_4545|M.abscessus_ATCC_1997 ------------MSAPHKPDSLRASGSSQPDIKPRSRDVTDGLEKTAARG
MSMEG_0482|M.smegmatis_MC2_155 --------------------MTDNFERAGAPGELRSNFTPGSTRWAVRRA
: **. .... . :. *.
MMAR_0432|M.marinum_M MLRAVGMVDEDFAKAQIGVASSWNEITPCNLSLDRLAKSVKEGVFAAGGY
MUL_1082|M.ulcerans_Agy99 MLRAVGMVDEDFAKAQIGVASSWNEITPCNLSLDRLAKSVKEGVFAAGGY
Mb0195c|M.bovis_AF2122/97 MLRAVGMDDEDFAKPQIGVASSWNEITPCNLSLDRLANAVKEGVFSAGGY
Rv0189c|M.tuberculosis_H37Rv MLRAVGMDDEDFAKPQIGVASSWNEITPCNLSLDRLANAVKEGVFSAGGY
MLBr_02608|M.leprae_Br4923 MLRAVGMNDDDFAKAQIGVASSWNEITPCNLSLDRLAKAVKEGVFSAGGY
MAV_4989|M.avium_104 MLRAVGMGDEDFAKPQIGVASSWNEITPCNLSLDRLAKAVKEGVFAAGGY
TH_0983|M.thermoresistible__bu MLRAVGMGDEDFAKPQIGVASSWNEITPCNLSLDRLAKAVKDGVHAAGGY
Mvan_0173|M.vanbaalenii_PYR-1 MLRAVGMGDEDFAKPQIGVGSSWNEITPCNLSLDRLAKAVKEGVFEAGGF
Mflv_0486|M.gilvum_PYR-GCK MLRAVGMTDDDWVKPQIGVGSSWNEITPCNMSLQRLAQAVKGGVHEAGGY
MAB_4545|M.abscessus_ATCC_1997 MLRAVGMGDDDWVKPQIGVGSSWNEITPCNMSLQRLAHSVKDGVHEAGGY
MSMEG_0482|M.smegmatis_MC2_155 QWSAMGIDWRDQEKPKIAIVNTSSKLSVCFAHLDDVAVRVAEAVRAAGGL
*:*: * *.:*.: .: .::: * *: :* * .* ***
MMAR_0432|M.marinum_M PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
MUL_1082|M.ulcerans_Agy99 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
Mb0195c|M.bovis_AF2122/97 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVEVVMQAERLDGSVLLA
Rv0189c|M.tuberculosis_H37Rv PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVEVVMQAERLDGSVLLA
MLBr_02608|M.leprae_Br4923 PLEFGTISVSDGISMGHQGMHFSLVSREVIADSVETVMQAERLDGSVLLA
MAV_4989|M.avium_104 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
TH_0983|M.thermoresistible__bu PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
Mvan_0173|M.vanbaalenii_PYR-1 PMEFGTISVSDGISMGHEGMHFSLVSREIIADSVETVMQAERLDGSVLLA
Mflv_0486|M.gilvum_PYR-GCK PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVVQAERLDGTVLLA
MAB_4545|M.abscessus_ATCC_1997 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
MSMEG_0482|M.smegmatis_MC2_155 PFEIRTTAPSDFVTSAGRKARYLMPTRDLIVNDVEAAVEGAVLDGVVFLS
*:*: * : ** :: . . :: : :*::*.:.**..::. *** *:*:
MMAR_0432|M.marinum_M GCDKSLPGMLMAAARLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFE
MUL_1082|M.ulcerans_Agy99 GCDKSLPGMLMAAARLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFE
Mb0195c|M.bovis_AF2122/97 GCDKSLPGMLMAAARLDLAAVFLYAGSILPGRAKLSDGSERDVTIIDAFE
Rv0189c|M.tuberculosis_H37Rv GCDKSLPGMLMAAARLDLAAVFLYAGSILPGRAKLSDGSERDVTIIDAFE
MLBr_02608|M.leprae_Br4923 GCDKSLPGMLMAAARLDLASVFLYAGSILPGRTKLSDGTEHEVTLIDAFE
MAV_4989|M.avium_104 GCDKSLPGMLMAAARLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFE
TH_0983|M.thermoresistible__bu GCDKSLPGMLMAAARLDLAAVFLYAGSILPGIAKLSDGTEREVTIIDAFE
Mvan_0173|M.vanbaalenii_PYR-1 GCDKSLPGMLMAAARLDLASVFLYAGSILPGRVKLSDGSEKDVTIIDAFE
Mflv_0486|M.gilvum_PYR-GCK GCDKSIPGMLMAAARLDLASVFFYNGSIMPGVAKLTDGSEKEVTIIDAFE
MAB_4545|M.abscessus_ATCC_1997 GCDKSIPGMLMAAARLDLASVFLYNGSIMPGKAKLTDGTEKEVTIIDAFE
MSMEG_0482|M.smegmatis_MC2_155 SCDKTTPAHLMAAARLDLPAIVVIG-----GYQRGGTWSGCSVDIDTVYE
.***: *. *********.::.. * : : .* : .:*
MMAR_0432|M.marinum_M AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
MUL_1082|M.ulcerans_Agy99 AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
Mb0195c|M.bovis_AF2122/97 AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
Rv0189c|M.tuberculosis_H37Rv AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
MLBr_02608|M.leprae_Br4923 AVGACSRGLMPRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
MAV_4989|M.avium_104 AVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
TH_0983|M.thermoresistible__bu AVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
Mvan_0173|M.vanbaalenii_PYR-1 AVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
Mflv_0486|M.gilvum_PYR-GCK AVGACARGLMSREDVDIIERAICPGEGACGGMYTANTMASAAEALGMSLP
MAB_4545|M.abscessus_ATCC_1997 AVGACARGLMSREDVDIIERAICPGEGACGGMYTANTMASAAEALGMSLP
MSMEG_0482|M.smegmatis_MC2_155 SVGALTAGTMTVDQLGELADQAIEGPGVCAGLATANTMHCLAEALGMTVA
:*** : * *. ::. : * *.*.*: ***** . ******::.
MMAR_0432|M.marinum_M GSAAPPATDRRRDGFARQSGQAVIELLRQGITARDIMTREAFENAIAVVM
MUL_1082|M.ulcerans_Agy99 GSAAPPATDRRRDGFARQSGQAVIELLRQGITARDIMTREAFENAIAVVM
Mb0195c|M.bovis_AF2122/97 GSAAPPATDRRRDGFARRSGQAVVELLRRGITARDILTKEAFENAIAVVM
Rv0189c|M.tuberculosis_H37Rv GSAAPPATDRRRDGFARRSGQAVVELLRRGITARDILTKEAFENAIAVVM
MLBr_02608|M.leprae_Br4923 GSAAPPATDRRRDGFARRSGQAVIELLRRGITARDILTKEAFENAIAVVM
MAV_4989|M.avium_104 GSAAPPATDRRRDGFARRSGQAVVELLRRGITARDILTKEAFENAIAVVM
TH_0983|M.thermoresistible__bu GSAAPPATDRRRDGFARKSGQAVVELLRRGITARDILTKEAFENAIAVVM
Mvan_0173|M.vanbaalenii_PYR-1 GSAAPPATDRRRDGFARKSGMAVVELLRRGITARDILTKEAFENAIAVVM
Mflv_0486|M.gilvum_PYR-GCK GSASPVAIDKRRDDFARRSGEAVVEMLRRGITARDILTKEAFENAIAVVM
MAB_4545|M.abscessus_ATCC_1997 GSASPVAIDKRREEYARKSGEAVVEMLRRGITARDILTKEAFENAIAVVM
MSMEG_0482|M.smegmatis_MC2_155 GSAPVRANSDRMLDNAAAAGHHIVKMVEQNLTARQVITEKSIENAIEVAL
***. * . * * :* :::::.:.:***:::*.:::**** *.:
MMAR_0432|M.marinum_M AFGGSTNAVLHLLAIAHEANVELTLD-DFSRIGSKVPHLADVKPFGRHVM
MUL_1082|M.ulcerans_Agy99 AFGGSTNAVLHLLAIAHEANVELTLD-DFSRIGSKVPHLADVKPFGRHVM
Mb0195c|M.bovis_AF2122/97 AFGGSTNAVLHLLAIAHEANVALSLQ-DFSRIGSGVPHLADVKPFGRHVM
Rv0189c|M.tuberculosis_H37Rv AFGGSTNAVLHLLAIAHEANVALSLQ-DFSRIGSGVPHLADVKPFGRHVM
MLBr_02608|M.leprae_Br4923 AFGGSTNAILHLLAIAHEADVTLSLE-DFSRIGFKVPHIADVKPFGRHVM
MAV_4989|M.avium_104 AFGGSTNAVLHLLAIAHEADVALSLD-DFSRIGSKVPHLADVKPFGRHVM
TH_0983|M.thermoresistible__bu AFGGSTNAVLHLLAIAHEANVKLTLE-DFTRIGNKVPHLADVKPFGAHVM
Mvan_0173|M.vanbaalenii_PYR-1 AFGGSTNAVLHLLAIAYEANVKLTLE-DFTRVGAKVPHLADVKPFGAYVM
Mflv_0486|M.gilvum_PYR-GCK AFGGSTNAVLHLLAIAWEAQVELTLA-DFTRIGQKVPHLADVKPFGAYVM
MAB_4545|M.abscessus_ATCC_1997 AFGGSTNAVLHLLAIAKEAEVELSLA-DFTRIGNKVPHLADVKPFGRHVM
MSMEG_0482|M.smegmatis_MC2_155 ALGGSVNCIRHLAAICAEADLDLDVVGMFEKHGGDAVQLAAIRPNGAHQV
*:***.*.: ** **. **:: * : * : * . ::* ::* * : :
MMAR_0432|M.marinum_M SHVDHIGGVPVVMKTLLDAGLLHGDCLTVTGRTVAENLEAIAPPDPDGKV
MUL_1082|M.ulcerans_Agy99 SHVDHIGGVPVVMKTLLDAGLLHGDCLTVTGRTVAENLEAIAPPDPDGKV
Mb0195c|M.bovis_AF2122/97 SDVDHIGGVPVVMKALLDAGLLHGDCLTVTGHTMAENLAAITPPDPDGKV
Rv0189c|M.tuberculosis_H37Rv SDVDHIGGVPVVMKALLDAGLLHGDCLTVTGHTMAENLAAITPPDPDGKV
MLBr_02608|M.leprae_Br4923 FDVDHIGGVPVVMKALLDAGLLHGDCLTVTGQTMAENLASIAPPDPDGQV
MAV_4989|M.avium_104 TDVDHIGGVPVMMKALLDAGLLNGDCLTVTGATVAQNLAAIAPPDPDGKV
TH_0983|M.thermoresistible__bu FDVDHIGGVPVIMKTLLDAGLLHGDCLTVTGKTMAENLAHIAPPDPDGKV
Mvan_0173|M.vanbaalenii_PYR-1 NDVDRIGGVPVVMKALLDAGLLHGDCLTVTGKTMAENLAHIAPPDPDGKV
Mflv_0486|M.gilvum_PYR-GCK KHVDEIGGVPVVMRALLDAGLLHGDCLTVTGQTMAENLAHIEPPDPDGKV
MAB_4545|M.abscessus_ATCC_1997 KDVDEIGGVPVVMRALLDAGLLHGDCLTVTGKTMAENLAHIAPPDPDGKV
MSMEG_0482|M.smegmatis_MC2_155 WDLDGVGGIQTVMRTLLPR--LHGDALTVDGRTVSERAADAQP--ADGDV
.:* :**: .:*::** *:**.*** * *:::. * .**.*
MMAR_0432|M.marinum_M LRALNNPIHATGGITILRGSLAPEGAVVKTAGLDTDV---FEGTARVFDG
MUL_1082|M.ulcerans_Agy99 LRALINPIHATGGITILRGSLAPEGAVVKTAGLDTDV---FEGTARVFDG
Mb0195c|M.bovis_AF2122/97 LRALANPIHPSGGITILHGSLAPEGAVVKTAGFDSDV---FEGTARVFDG
Rv0189c|M.tuberculosis_H37Rv LRALANPIHPSGGITILHGSLAPEGAVVKTAGFDSDV---FEGTARVFDG
MLBr_02608|M.leprae_Br4923 IRTLHNPIHPTGGITILRGSLAPDGAVVKTAGLDSDV---FEGTARVFDG
MAV_4989|M.avium_104 LRALSDPLHPTGGITILRGSLAPEGAVVKSAGFDSDV---FEGTARVFDG
TH_0983|M.thermoresistible__bu LRALSNPIHPTGGITILHGSLAPEGAVVKSAGFDTSV---FEGTARVFDR
Mvan_0173|M.vanbaalenii_PYR-1 LRALSRPIHPTGGITILHGSLAPEGAVVKSAGFDSDV---FEGTARVFER
Mflv_0486|M.gilvum_PYR-GCK LRAMNNPIHPTGGITILHGSLAPEGAVVKSAGFDSDV---FEGTARVFER
MAB_4545|M.abscessus_ATCC_1997 LRAMNDPIHPTGGITILHGSLAPEGAVVKSAGFESDV---FEGTARVFER
MSMEG_0482|M.smegmatis_MC2_155 LHPLDDPINDEPGLIILRGSLAPDGSIVKVAGVGKATRRQFNGPAKVFVG
::.: *:: *: **:*****:*::** **. . . *:*.*:**
MMAR_0432|M.marinum_M ERAALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
MUL_1082|M.ulcerans_Agy99 ERAALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
Mb0195c|M.bovis_AF2122/97 ERAALDALEDGTITVGDAVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
Rv0189c|M.tuberculosis_H37Rv ERAALDALEDGTITVGDAVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
MLBr_02608|M.leprae_Br4923 ERAALDALKDGTIAKGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
MAV_4989|M.avium_104 ERAALDALEDGTITKGDAVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
TH_0983|M.thermoresistible__bu ERAAMDALEDGTIQAGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
Mvan_0173|M.vanbaalenii_PYR-1 ERAALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
Mflv_0486|M.gilvum_PYR-GCK ERAALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
MAB_4545|M.abscessus_ATCC_1997 ERAALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKD
MSMEG_0482|M.smegmatis_MC2_155 EDDAIAALGDGRIQRGDVIVLRGMGPRGGPGTVFAASFVAALNGAGLGGH
* *: ** ** * **.:*:* **:**** ::..*::***** .
MMAR_0432|M.marinum_M VLLLTDGRFSGGTTGFCVGHIAPEAVDAGPIAFVRDGDRIRLDVAGRTLD
MUL_1082|M.ulcerans_Agy99 VLLLTDGRFSGGTTGFCVGHIAPEAVDAGPIAFVRDGDRIRLDVAGRTLD
Mb0195c|M.bovis_AF2122/97 VLLLTDGRFSGGTTGLCVGHIAPEAVDGGPIALLRNGDRIRLDVAGRVLD
Rv0189c|M.tuberculosis_H37Rv VLLLTDGRFSGGTTGLCVGHIAPEAVDGGPIALLRNGDRIRLDVAGRVLD
MLBr_02608|M.leprae_Br4923 VLLLTDGRFSGGTTGFCVGHIAPEAVEAGPIAFLRDGDRVLLDVVGCSLD
MAV_4989|M.avium_104 VLLLTDGRFSGGTTGLCVGHIAPEAVDAGPIAFLRDGDRIRLDVANRVLD
TH_0983|M.thermoresistible__bu VLLLTDGRFSGGTTGLCVGHVAPEAVDGGPIAFLRDGDRIRLDVAKGTLD
Mvan_0173|M.vanbaalenii_PYR-1 VLLMTDGRFSGGTTGLCVGHIAPEAVDGGPIAFIKDGDKIRLDVAKGTLD
Mflv_0486|M.gilvum_PYR-GCK VLLMTDGRFSGGTTGLCVGHIAPEAVDGGPIAFVRDGDRIRLDVANGTLD
MAB_4545|M.abscessus_ATCC_1997 VLLMTDGRFSGGTTGLCVGHIAPEAVDGGPIAFVRDGDRIRLDVANGKLD
MSMEG_0482|M.smegmatis_MC2_155 VAVVTDGELSGLNHGLVVGQVMPEAADGGPLAGLQNGDTVAIDLDARRID
* ::***.:** . *: **:: ***.:.**:* :::** : :*: :*
MMAR_0432|M.marinum_M VLTDPAEFAARQQNFTPPAPRYKTGVLAKYVKLVGSAAIGAVCG----
MUL_1082|M.ulcerans_Agy99 VLTDPPEFAARQQNFTPPAPRYKTGVLAKYVKLVGSAAIGAVCG----
Mb0195c|M.bovis_AF2122/97 VLADPAEFASRQQDFSPPPPRYTTGVLSKYVKLVSSAAVGAVCG----
Rv0189c|M.tuberculosis_H37Rv VLADPAEFASRQQDFSPPPPRYTTGVLSKYVKLVSSAAVGAVCG----
MLBr_02608|M.leprae_Br4923 VLVDPVEFSSRKKDFIPPAPRYTTGVLAKYVKLVSSATVGAVCG----
MAV_4989|M.avium_104 VLVDPAEFDSRRTAFTPPPPRYKTGVLAKYVKLVGSAAIGAVCG----
TH_0983|M.thermoresistible__bu VLVDPAEFEARKKDWEPLPPRYTTGVLAKYTKLVGSAATGAVCT----
Mvan_0173|M.vanbaalenii_PYR-1 VVVDKDEFEARKVGFEPLPPVYTTGVLAKYTKLVGSAATGAVCT----
Mflv_0486|M.gilvum_PYR-GCK ILVDEAEFESRKAGFEPLPPVYTTGVLAKYTKLVGSAAVGAVCS----
MAB_4545|M.abscessus_ATCC_1997 LLVDEAEIAERRKGFEPLPPRYKTGVLAKYTKLVQSAAVGAVCG----
MSMEG_0482|M.smegmatis_MC2_155 SHPIRDGEPVRATGEHP-----ERGWLGQYAALVGPVQRGAVLRRPRN
* * * *.:*. ** .. ***