For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAPHKPDSLRASGSSQPDIKPRSRDVTDGLEKTAARGMLRAVGMGDDDWVKPQIGVGSSWNEITPCNMS LQRLAHSVKDGVHEAGGYPLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLAGC DKSIPGMLMAAARLDLASVFLYNGSIMPGKAKLTDGTEKEVTIIDAFEAVGACARGLMSREDVDIIERAI CPGEGACGGMYTANTMASAAEALGMSLPGSASPVAIDKRREEYARKSGEAVVEMLRRGITARDILTKEAF ENAIAVVMAFGGSTNAVLHLLAIAKEAEVELSLADFTRIGNKVPHLADVKPFGRHVMKDVDEIGGVPVVM RALLDAGLLHGDCLTVTGKTMAENLAHIAPPDPDGKVLRAMNDPIHPTGGITILHGSLAPEGAVVKSAGF ESDVFEGTARVFERERAALDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLMTD GRFSGGTTGLCVGHIAPEAVDGGPIAFVRDGDRIRLDVANGKLDLLVDEAEIAERRKGFEPLPPRYKTGV LAKYTKLVQSAAVGAVCG
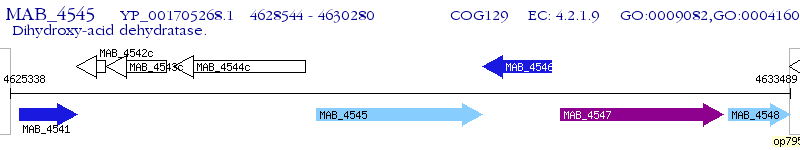
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4545 | - | - | 100% (578) | dihydroxy-acid dehydratase |
| M. abscessus ATCC 19977 | MAB_4640c | - | e-114 | 40.47% (593) | dihydroxy-acid dehydratase (IlvD) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0195c | ilvD | 0.0 | 85.19% (567) | dihydroxy-acid dehydratase |
| M. gilvum PYR-GCK | Mflv_0486 | - | 0.0 | 92.37% (577) | dihydroxy-acid dehydratase |
| M. tuberculosis H37Rv | Rv0189c | ilvD | 0.0 | 85.19% (567) | dihydroxy-acid dehydratase |
| M. leprae Br4923 | MLBr_02608 | ilvD | 0.0 | 83.60% (561) | dihydroxy-acid dehydratase |
| M. marinum M | MMAR_0432 | ilvD | 0.0 | 85.05% (562) | dihydroxy-acid dehydratase IlvD |
| M. avium 104 | MAV_4989 | ilvD | 0.0 | 86.51% (571) | dihydroxy-acid dehydratase |
| M. smegmatis MC2 155 | MSMEG_0229 | ilvD | 0.0 | 93.39% (560) | dihydroxy-acid dehydratase |
| M. thermoresistible (build 8) | TH_0983 | ilvD | 0.0 | 88.26% (562) | PROBABLE DIHYDROXY-ACID DEHYDRATASE ILVD (DAD) |
| M. ulcerans Agy99 | MUL_1082 | ilvD | 0.0 | 84.70% (562) | dihydroxy-acid dehydratase |
| M. vanbaalenii PYR-1 | Mvan_0173 | - | 0.0 | 88.08% (562) | dihydroxy-acid dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0432|M.marinum_M --------------------------MPEADIKPRSRDVTDGLEKAAARG
MUL_1082|M.ulcerans_Agy99 --------------------------MPEADIKPRSRDVTDGLEKAAARG
Mb0195c|M.bovis_AF2122/97 ---------------MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARG
Rv0189c|M.tuberculosis_H37Rv ---------------MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARG
MLBr_02608|M.leprae_Br4923 --------------------------MMPPDIKPRSRDVTDGLEKAAARG
MAV_4989|M.avium_104 ---------------MPTTDSARAADIKQPDIKPRSRDVTDGLEKAAARG
TH_0983|M.thermoresistible__bu ------------------------MAQNQPDIKPRSRDVTDGLEKAAARG
Mvan_0173|M.vanbaalenii_PYR-1 -------------------------MPSSPDIKPRSRDVTDGLEKAAARG
MAB_4545|M.abscessus_ATCC_1997 ------------MSAPHKPDSLRASGSSQPDIKPRSRDVTDGLEKTAARG
Mflv_0486|M.gilvum_PYR-GCK MWPMNRRAHNHPMPSDSRSHSLRASGSS-VDIKPRSRDVTDGLERTAARG
MSMEG_0229|M.smegmatis_MC2_155 ------------MPSDDKS-------TETPDIKPRSRDVTDGLEKAAARG
**************::****
MMAR_0432|M.marinum_M MLRAVGMVDEDFAKAQIGVASSWNEITPCNLSLDRLAKSVKEGVFAAGGY
MUL_1082|M.ulcerans_Agy99 MLRAVGMVDEDFAKAQIGVASSWNEITPCNLSLDRLAKSVKEGVFAAGGY
Mb0195c|M.bovis_AF2122/97 MLRAVGMDDEDFAKPQIGVASSWNEITPCNLSLDRLANAVKEGVFSAGGY
Rv0189c|M.tuberculosis_H37Rv MLRAVGMDDEDFAKPQIGVASSWNEITPCNLSLDRLANAVKEGVFSAGGY
MLBr_02608|M.leprae_Br4923 MLRAVGMNDDDFAKAQIGVASSWNEITPCNLSLDRLAKAVKEGVFSAGGY
MAV_4989|M.avium_104 MLRAVGMGDEDFAKPQIGVASSWNEITPCNLSLDRLAKAVKEGVFAAGGY
TH_0983|M.thermoresistible__bu MLRAVGMGDEDFAKPQIGVASSWNEITPCNLSLDRLAKAVKDGVHAAGGY
Mvan_0173|M.vanbaalenii_PYR-1 MLRAVGMGDEDFAKPQIGVGSSWNEITPCNLSLDRLAKAVKEGVFEAGGF
MAB_4545|M.abscessus_ATCC_1997 MLRAVGMGDDDWVKPQIGVGSSWNEITPCNMSLQRLAHSVKDGVHEAGGY
Mflv_0486|M.gilvum_PYR-GCK MLRAVGMTDDDWVKPQIGVGSSWNEITPCNMSLQRLAQAVKGGVHEAGGY
MSMEG_0229|M.smegmatis_MC2_155 MLRAVGMGDADWAKPQIGVGSSWNEITPCNMSLQRLAKSVKDGVHEAGGY
******* * *:.*.****.**********:**:***::** **. ***:
MMAR_0432|M.marinum_M PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
MUL_1082|M.ulcerans_Agy99 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
Mb0195c|M.bovis_AF2122/97 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVEVVMQAERLDGSVLLA
Rv0189c|M.tuberculosis_H37Rv PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVEVVMQAERLDGSVLLA
MLBr_02608|M.leprae_Br4923 PLEFGTISVSDGISMGHQGMHFSLVSREVIADSVETVMQAERLDGSVLLA
MAV_4989|M.avium_104 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
TH_0983|M.thermoresistible__bu PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
Mvan_0173|M.vanbaalenii_PYR-1 PMEFGTISVSDGISMGHEGMHFSLVSREIIADSVETVMQAERLDGSVLLA
MAB_4545|M.abscessus_ATCC_1997 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLA
Mflv_0486|M.gilvum_PYR-GCK PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVVQAERLDGTVLLA
MSMEG_0229|M.smegmatis_MC2_155 PLEFGTISVSDGISMGHEGMHFSLVSREVIADSVETVVQAERLDGTVLLA
*:***************:**********:******.*:*******:****
MMAR_0432|M.marinum_M GCDKSLPGMLMAAARLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFE
MUL_1082|M.ulcerans_Agy99 GCDKSLPGMLMAAARLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFE
Mb0195c|M.bovis_AF2122/97 GCDKSLPGMLMAAARLDLAAVFLYAGSILPGRAKLSDGSERDVTIIDAFE
Rv0189c|M.tuberculosis_H37Rv GCDKSLPGMLMAAARLDLAAVFLYAGSILPGRAKLSDGSERDVTIIDAFE
MLBr_02608|M.leprae_Br4923 GCDKSLPGMLMAAARLDLASVFLYAGSILPGRTKLSDGTEHEVTLIDAFE
MAV_4989|M.avium_104 GCDKSLPGMLMAAARLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFE
TH_0983|M.thermoresistible__bu GCDKSLPGMLMAAARLDLAAVFLYAGSILPGIAKLSDGTEREVTIIDAFE
Mvan_0173|M.vanbaalenii_PYR-1 GCDKSLPGMLMAAARLDLASVFLYAGSILPGRVKLSDGSEKDVTIIDAFE
MAB_4545|M.abscessus_ATCC_1997 GCDKSIPGMLMAAARLDLASVFLYNGSIMPGKAKLTDGTEKEVTIIDAFE
Mflv_0486|M.gilvum_PYR-GCK GCDKSIPGMLMAAARLDLASVFFYNGSIMPGVAKLTDGSEKEVTIIDAFE
MSMEG_0229|M.smegmatis_MC2_155 GCDKSIPGMLMAAARLDLASVFFYNGSIMPGVAKLTDGTEKEVTIIDAFE
*****:*************:**:* ***:** .**:**:*::**:*****
MMAR_0432|M.marinum_M AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
MUL_1082|M.ulcerans_Agy99 AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
Mb0195c|M.bovis_AF2122/97 AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
Rv0189c|M.tuberculosis_H37Rv AVGACSRGLMSRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
MLBr_02608|M.leprae_Br4923 AVGACSRGLMPRADVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
MAV_4989|M.avium_104 AVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
TH_0983|M.thermoresistible__bu AVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
Mvan_0173|M.vanbaalenii_PYR-1 AVGACARGLMPREDVDAIERAICPGEGACGGMYTANTMASAAEALGMSLP
MAB_4545|M.abscessus_ATCC_1997 AVGACARGLMSREDVDIIERAICPGEGACGGMYTANTMASAAEALGMSLP
Mflv_0486|M.gilvum_PYR-GCK AVGACARGLMSREDVDIIERAICPGEGACGGMYTANTMASAAEALGMSLP
MSMEG_0229|M.smegmatis_MC2_155 AVGACVRGLMSREDVDIIERAICPGEGACGGMYTANTMASAAEALGLSLP
***** ****.* *** *****************************:***
MMAR_0432|M.marinum_M GSAAPPATDRRRDGFARQSGQAVIELLRQGITARDIMTREAFENAIAVVM
MUL_1082|M.ulcerans_Agy99 GSAAPPATDRRRDGFARQSGQAVIELLRQGITARDIMTREAFENAIAVVM
Mb0195c|M.bovis_AF2122/97 GSAAPPATDRRRDGFARRSGQAVVELLRRGITARDILTKEAFENAIAVVM
Rv0189c|M.tuberculosis_H37Rv GSAAPPATDRRRDGFARRSGQAVVELLRRGITARDILTKEAFENAIAVVM
MLBr_02608|M.leprae_Br4923 GSAAPPATDRRRDGFARRSGQAVIELLRRGITARDILTKEAFENAIAVVM
MAV_4989|M.avium_104 GSAAPPATDRRRDGFARRSGQAVVELLRRGITARDILTKEAFENAIAVVM
TH_0983|M.thermoresistible__bu GSAAPPATDRRRDGFARKSGQAVVELLRRGITARDILTKEAFENAIAVVM
Mvan_0173|M.vanbaalenii_PYR-1 GSAAPPATDRRRDGFARKSGMAVVELLRRGITARDILTKEAFENAIAVVM
MAB_4545|M.abscessus_ATCC_1997 GSASPVAIDKRREEYARKSGEAVVEMLRRGITARDILTKEAFENAIAVVM
Mflv_0486|M.gilvum_PYR-GCK GSASPVAIDKRRDDFARRSGEAVVEMLRRGITARDILTKEAFENAIAVVM
MSMEG_0229|M.smegmatis_MC2_155 GSASPVAIDKRRDEFARRSGEAVVDMLRRGITARDILTKEAFENAIAVVM
***:* * *:**: :**:** **:::**:*******:*:***********
MMAR_0432|M.marinum_M AFGGSTNAVLHLLAIAHEANVELTLDDFSRIGSKVPHLADVKPFGRHVMS
MUL_1082|M.ulcerans_Agy99 AFGGSTNAVLHLLAIAHEANVELTLDDFSRIGSKVPHLADVKPFGRHVMS
Mb0195c|M.bovis_AF2122/97 AFGGSTNAVLHLLAIAHEANVALSLQDFSRIGSGVPHLADVKPFGRHVMS
Rv0189c|M.tuberculosis_H37Rv AFGGSTNAVLHLLAIAHEANVALSLQDFSRIGSGVPHLADVKPFGRHVMS
MLBr_02608|M.leprae_Br4923 AFGGSTNAILHLLAIAHEADVTLSLEDFSRIGFKVPHIADVKPFGRHVMF
MAV_4989|M.avium_104 AFGGSTNAVLHLLAIAHEADVALSLDDFSRIGSKVPHLADVKPFGRHVMT
TH_0983|M.thermoresistible__bu AFGGSTNAVLHLLAIAHEANVKLTLEDFTRIGNKVPHLADVKPFGAHVMF
Mvan_0173|M.vanbaalenii_PYR-1 AFGGSTNAVLHLLAIAYEANVKLTLEDFTRVGAKVPHLADVKPFGAYVMN
MAB_4545|M.abscessus_ATCC_1997 AFGGSTNAVLHLLAIAKEAEVELSLADFTRIGNKVPHLADVKPFGRHVMK
Mflv_0486|M.gilvum_PYR-GCK AFGGSTNAVLHLLAIAWEAQVELTLADFTRIGQKVPHLADVKPFGAYVMK
MSMEG_0229|M.smegmatis_MC2_155 AFGGSTNAVLHLLAIAREAEVELTLDDFTRVGSKVPHLADVKPFGRHVMK
********:******* **:* *:* **:*:* ***:******* :**
MMAR_0432|M.marinum_M HVDHIGGVPVVMKTLLDAGLLHGDCLTVTGRTVAENLEAIAPPDPDGKVL
MUL_1082|M.ulcerans_Agy99 HVDHIGGVPVVMKTLLDAGLLHGDCLTVTGRTVAENLEAIAPPDPDGKVL
Mb0195c|M.bovis_AF2122/97 DVDHIGGVPVVMKALLDAGLLHGDCLTVTGHTMAENLAAITPPDPDGKVL
Rv0189c|M.tuberculosis_H37Rv DVDHIGGVPVVMKALLDAGLLHGDCLTVTGHTMAENLAAITPPDPDGKVL
MLBr_02608|M.leprae_Br4923 DVDHIGGVPVVMKALLDAGLLHGDCLTVTGQTMAENLASIAPPDPDGQVI
MAV_4989|M.avium_104 DVDHIGGVPVMMKALLDAGLLNGDCLTVTGATVAQNLAAIAPPDPDGKVL
TH_0983|M.thermoresistible__bu DVDHIGGVPVIMKTLLDAGLLHGDCLTVTGKTMAENLAHIAPPDPDGKVL
Mvan_0173|M.vanbaalenii_PYR-1 DVDRIGGVPVVMKALLDAGLLHGDCLTVTGKTMAENLAHIAPPDPDGKVL
MAB_4545|M.abscessus_ATCC_1997 DVDEIGGVPVVMRALLDAGLLHGDCLTVTGKTMAENLAHIAPPDPDGKVL
Mflv_0486|M.gilvum_PYR-GCK HVDEIGGVPVVMRALLDAGLLHGDCLTVTGQTMAENLAHIEPPDPDGKVL
MSMEG_0229|M.smegmatis_MC2_155 HVDEIGGVPVVMRALLDAGLLNGECLTVTGKTMAENLAHIAPPDPDGKVL
.**.******:*::*******:*:****** *:*:** * ******:*:
MMAR_0432|M.marinum_M RALNNPIHATGGITILRGSLAPEGAVVKTAGLDTDVFEGTARVFDGERAA
MUL_1082|M.ulcerans_Agy99 RALINPIHATGGITILRGSLAPEGAVVKTAGLDTDVFEGTARVFDGERAA
Mb0195c|M.bovis_AF2122/97 RALANPIHPSGGITILHGSLAPEGAVVKTAGFDSDVFEGTARVFDGERAA
Rv0189c|M.tuberculosis_H37Rv RALANPIHPSGGITILHGSLAPEGAVVKTAGFDSDVFEGTARVFDGERAA
MLBr_02608|M.leprae_Br4923 RTLHNPIHPTGGITILRGSLAPDGAVVKTAGLDSDVFEGTARVFDGERAA
MAV_4989|M.avium_104 RALSDPLHPTGGITILRGSLAPEGAVVKSAGFDSDVFEGTARVFDGERAA
TH_0983|M.thermoresistible__bu RALSNPIHPTGGITILHGSLAPEGAVVKSAGFDTSVFEGTARVFDRERAA
Mvan_0173|M.vanbaalenii_PYR-1 RALSRPIHPTGGITILHGSLAPEGAVVKSAGFDSDVFEGTARVFERERAA
MAB_4545|M.abscessus_ATCC_1997 RAMNDPIHPTGGITILHGSLAPEGAVVKSAGFESDVFEGTARVFERERAA
Mflv_0486|M.gilvum_PYR-GCK RAMNNPIHPTGGITILHGSLAPEGAVVKSAGFDSDVFEGTARVFERERAA
MSMEG_0229|M.smegmatis_MC2_155 RAMNNPIHPTGGITILHGSLAPEGAVVKSAGFDSDVFEGTARVFDRERAA
*:: *:*.:******:*****:*****:**:::.*********: ****
MMAR_0432|M.marinum_M LDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLL
MUL_1082|M.ulcerans_Agy99 LDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLL
Mb0195c|M.bovis_AF2122/97 LDALEDGTITVGDAVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLL
Rv0189c|M.tuberculosis_H37Rv LDALEDGTITVGDAVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLL
MLBr_02608|M.leprae_Br4923 LDALKDGTIAKGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLL
MAV_4989|M.avium_104 LDALEDGTITKGDAVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLL
TH_0983|M.thermoresistible__bu MDALEDGTIQAGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLL
Mvan_0173|M.vanbaalenii_PYR-1 LDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLM
MAB_4545|M.abscessus_ATCC_1997 LDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLM
Mflv_0486|M.gilvum_PYR-GCK LDALEDGTITHGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLM
MSMEG_0229|M.smegmatis_MC2_155 MDALEDGTIQAGDVVVIRYEGPKGGPGMREMLAITGAIKGAGLGKDVLLM
:***:**** **.***********************************:
MMAR_0432|M.marinum_M TDGRFSGGTTGFCVGHIAPEAVDAGPIAFVRDGDRIRLDVAGRTLDVLTD
MUL_1082|M.ulcerans_Agy99 TDGRFSGGTTGFCVGHIAPEAVDAGPIAFVRDGDRIRLDVAGRTLDVLTD
Mb0195c|M.bovis_AF2122/97 TDGRFSGGTTGLCVGHIAPEAVDGGPIALLRNGDRIRLDVAGRVLDVLAD
Rv0189c|M.tuberculosis_H37Rv TDGRFSGGTTGLCVGHIAPEAVDGGPIALLRNGDRIRLDVAGRVLDVLAD
MLBr_02608|M.leprae_Br4923 TDGRFSGGTTGFCVGHIAPEAVEAGPIAFLRDGDRVLLDVVGCSLDVLVD
MAV_4989|M.avium_104 TDGRFSGGTTGLCVGHIAPEAVDAGPIAFLRDGDRIRLDVANRVLDVLVD
TH_0983|M.thermoresistible__bu TDGRFSGGTTGLCVGHVAPEAVDGGPIAFLRDGDRIRLDVAKGTLDVLVD
Mvan_0173|M.vanbaalenii_PYR-1 TDGRFSGGTTGLCVGHIAPEAVDGGPIAFIKDGDKIRLDVAKGTLDVVVD
MAB_4545|M.abscessus_ATCC_1997 TDGRFSGGTTGLCVGHIAPEAVDGGPIAFVRDGDRIRLDVANGKLDLLVD
Mflv_0486|M.gilvum_PYR-GCK TDGRFSGGTTGLCVGHIAPEAVDGGPIAFVRDGDRIRLDVANGTLDILVD
MSMEG_0229|M.smegmatis_MC2_155 TDGRFSGGTTGLCVGHIAPEAVDGGPIAFVRDGDRIRLDVAKGTLDVLVD
***********:****:*****:.****::::**:: ***. **::.*
MMAR_0432|M.marinum_M PAEFAARQQNFTPPAPRYKTGVLAKYVKLVGSAAIGAVCG
MUL_1082|M.ulcerans_Agy99 PPEFAARQQNFTPPAPRYKTGVLAKYVKLVGSAAIGAVCG
Mb0195c|M.bovis_AF2122/97 PAEFASRQQDFSPPPPRYTTGVLSKYVKLVSSAAVGAVCG
Rv0189c|M.tuberculosis_H37Rv PAEFASRQQDFSPPPPRYTTGVLSKYVKLVSSAAVGAVCG
MLBr_02608|M.leprae_Br4923 PVEFSSRKKDFIPPAPRYTTGVLAKYVKLVSSATVGAVCG
MAV_4989|M.avium_104 PAEFDSRRTAFTPPPPRYKTGVLAKYVKLVGSAAIGAVCG
TH_0983|M.thermoresistible__bu PAEFEARKKDWEPLPPRYTTGVLAKYTKLVGSAATGAVCT
Mvan_0173|M.vanbaalenii_PYR-1 KDEFEARKVGFEPLPPVYTTGVLAKYTKLVGSAATGAVCT
MAB_4545|M.abscessus_ATCC_1997 EAEIAERRKGFEPLPPRYKTGVLAKYTKLVQSAAVGAVCG
Mflv_0486|M.gilvum_PYR-GCK EAEFESRKAGFEPLPPVYTTGVLAKYTKLVGSAAVGAVCS
MSMEG_0229|M.smegmatis_MC2_155 EEEFEARKAGFEPLPPRYKTGVLAKYTKLVQSAAVGAVCT
*: *: : * .* *.****:**.*** **: ****