For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASSSIPLRSRTVTHGRNMAGARALLRAAGVAREDFGKPIVAVANSFTEFVPGHTHLQPVGRIVSDAIRA AGGIPREFNTIAVDDGIAMGHSGMLYSLPSRDLIADSVEYMVQAHKADALVCISNCDKITPGMLMAALRL NVPTVFVSGGPMEGGRATLVDGRVRTGLHLIDSMSGAADPTTSDEDLARIEEAACPTCGSCSGMFTANSM NCLVEALGLALPGNGSLLATHTARKALYQNAGRTVMELAERYYDQGDVSALPRAVASRAAFENAMAMDIA MGGSTNTVLHLLAAAHEADLDFAITEIDEISRRIPCLCKVAPNGSYLMEDVHRAGGIPAILGELHRAGLL AKDVHAIHAATLDDWLDAWDIRAADPSDAAVDLFHAAPGGRRSATAFSQSERWESLDLDAMAGCVRDVDH AYSPDGGLAILTGNLAPDGAVVKTAGVDENLLTFTGPAVVVESQEEAVDAILGDRITAGDVLVVRYEGPR GGPGMQEMLYPTSFLKGRGLGKSCALITDGRFSGGSSGLSIGHVSPEAAAGGPIALIRDGDRVTIDIPSR TVTLDVDGDELARRRDELLASGGYRPRDRVRPVSAALRAYALLATSADRGAVRDVDNVVSDR
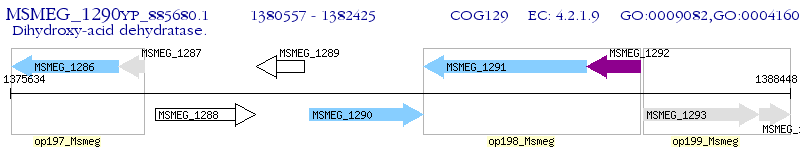
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1290 | ilvD | - | 100% (622) | dihydroxy-acid dehydratase |
| M. smegmatis MC2 155 | MSMEG_0229 | ilvD | e-117 | 40.43% (606) | dihydroxy-acid dehydratase |
| M. smegmatis MC2 155 | MSMEG_0482 | - | 2e-73 | 32.29% (607) | dihydroxy-acid dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0195c | ilvD | 1e-119 | 41.08% (611) | dihydroxy-acid dehydratase |
| M. gilvum PYR-GCK | Mflv_5171 | - | 0.0 | 77.30% (608) | dihydroxy-acid dehydratase |
| M. tuberculosis H37Rv | Rv0189c | ilvD | 1e-119 | 41.08% (611) | dihydroxy-acid dehydratase |
| M. leprae Br4923 | MLBr_02608 | ilvD | 1e-117 | 39.58% (614) | dihydroxy-acid dehydratase |
| M. abscessus ATCC 19977 | MAB_4640c | - | 0.0 | 75.38% (597) | dihydroxy-acid dehydratase (IlvD) |
| M. marinum M | MMAR_0432 | ilvD | 1e-117 | 40.55% (614) | dihydroxy-acid dehydratase IlvD |
| M. avium 104 | MAV_4989 | ilvD | 1e-119 | 41.58% (606) | dihydroxy-acid dehydratase |
| M. thermoresistible (build 8) | TH_0983 | ilvD | 1e-120 | 42.17% (607) | PROBABLE DIHYDROXY-ACID DEHYDRATASE ILVD (DAD) |
| M. ulcerans Agy99 | MUL_1082 | ilvD | 1e-117 | 40.55% (614) | dihydroxy-acid dehydratase |
| M. vanbaalenii PYR-1 | Mvan_1161 | - | 0.0 | 77.91% (611) | dihydroxy-acid dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0432|M.marinum_M -MP-----------EADIKPRSRDVTDGLEKAAARGMLRAVGMVDEDFAK
MUL_1082|M.ulcerans_Agy99 -MP-----------EADIKPRSRDVTDGLEKAAARGMLRAVGMVDEDFAK
MAV_4989|M.avium_104 -MPTTDSARAADIKQPDIKPRSRDVTDGLEKAAARGMLRAVGMGDEDFAK
Mb0195c|M.bovis_AF2122/97 -MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARGMLRAVGMDDEDFAK
Rv0189c|M.tuberculosis_H37Rv -MPQTTDEAASVSTVADIKPRSRDVTDGLEKAAARGMLRAVGMDDEDFAK
MLBr_02608|M.leprae_Br4923 MMP------------PDIKPRSRDVTDGLEKAAARGMLRAVGMNDDDFAK
TH_0983|M.thermoresistible__bu -MAQ---------NQPDIKPRSRDVTDGLEKAAARGMLRAVGMGDEDFAK
Mflv_5171|M.gilvum_PYR-GCK ----------------MPELRSRTVTHGRNMAGARALLRAAGVDGADIGK
Mvan_1161|M.vanbaalenii_PYR-1 -----------------MELRSRTVTHGRNMAGARSLLRAAGVDGADIGK
MSMEG_1290|M.smegmatis_MC2_155 ------------MASSSIPLRSRTVTHGRNMAGARALLRAAGVAREDFGK
MAB_4640c|M.abscessus_ATCC_199 ------------------------------MAGARALLRAAGVAASDFGK
*.**.:***.*: *:.*
MMAR_0432|M.marinum_M AQIGVASSWNEITPCNLSLDRLAKSVKEGVFAAGGYPLEFGTISVSDGIS
MUL_1082|M.ulcerans_Agy99 AQIGVASSWNEITPCNLSLDRLAKSVKEGVFAAGGYPLEFGTISVSDGIS
MAV_4989|M.avium_104 PQIGVASSWNEITPCNLSLDRLAKAVKEGVFAAGGYPLEFGTISVSDGIS
Mb0195c|M.bovis_AF2122/97 PQIGVASSWNEITPCNLSLDRLANAVKEGVFSAGGYPLEFGTISVSDGIS
Rv0189c|M.tuberculosis_H37Rv PQIGVASSWNEITPCNLSLDRLANAVKEGVFSAGGYPLEFGTISVSDGIS
MLBr_02608|M.leprae_Br4923 AQIGVASSWNEITPCNLSLDRLAKAVKEGVFSAGGYPLEFGTISVSDGIS
TH_0983|M.thermoresistible__bu PQIGVASSWNEITPCNLSLDRLAKAVKDGVHAAGGYPLEFGTISVSDGIS
Mflv_5171|M.gilvum_PYR-GCK PIVAVANSFTEFVPGHTHLQPVGRIVSEAIRAAGGVPREFNTIAVDDGIA
Mvan_1161|M.vanbaalenii_PYR-1 PIVAVANSFTEFVPGHTHLQPVGRIVSEAVTAAGGVPREFNTIAVDDGIA
MSMEG_1290|M.smegmatis_MC2_155 PIVAVANSFTEFVPGHTHLQPVGRIVSDAIRAAGGIPREFNTIAVDDGIA
MAB_4640c|M.abscessus_ATCC_199 PIVAVANSFAEFVPGHTHLQPVGRIVSEAIAAAGAVPREFNTIAVDDGIA
. :.**.*: *:.* : *: :.. *.:.: :**. * **.**:*.***:
MMAR_0432|M.marinum_M MGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLAGCDKSLPGMLMAAA
MUL_1082|M.ulcerans_Agy99 MGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLAGCDKSLPGMLMAAA
MAV_4989|M.avium_104 MGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLAGCDKSLPGMLMAAA
Mb0195c|M.bovis_AF2122/97 MGHEGMHFSLVSREVIADSVEVVMQAERLDGSVLLAGCDKSLPGMLMAAA
Rv0189c|M.tuberculosis_H37Rv MGHEGMHFSLVSREVIADSVEVVMQAERLDGSVLLAGCDKSLPGMLMAAA
MLBr_02608|M.leprae_Br4923 MGHQGMHFSLVSREVIADSVETVMQAERLDGSVLLAGCDKSLPGMLMAAA
TH_0983|M.thermoresistible__bu MGHEGMHFSLVSREVIADSVETVMQAERLDGSVLLAGCDKSLPGMLMAAA
Mflv_5171|M.gilvum_PYR-GCK MGHGGMLYSLPSRELIADSVEYMINAHCADAMVCISNCDKITPGMVMAAL
Mvan_1161|M.vanbaalenii_PYR-1 MGHGGMLYSLPSRDLIADSIEYMVNAHRADALVCISNCDKITPGMLMAAL
MSMEG_1290|M.smegmatis_MC2_155 MGHSGMLYSLPSRDLIADSVEYMVQAHKADALVCISNCDKITPGMLMAAL
MAB_4640c|M.abscessus_ATCC_199 MGHEGMLYSLPSRDLITDSIEYMVEAHRADALVCISNCDKITPGMLMAAL
*** ** :** **::*:**:* :::*. *. * ::.*** ***:***
MMAR_0432|M.marinum_M RLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFEAVGACSRGLMSRAD
MUL_1082|M.ulcerans_Agy99 RLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFEAVGACSRGLMSRAD
MAV_4989|M.avium_104 RLDLASVFLYAGSILPGVAKLSDGSEREVTIIDAFEAVGACARGLMPRED
Mb0195c|M.bovis_AF2122/97 RLDLAAVFLYAGSILPGRAKLSDGSERDVTIIDAFEAVGACSRGLMSRAD
Rv0189c|M.tuberculosis_H37Rv RLDLAAVFLYAGSILPGRAKLSDGSERDVTIIDAFEAVGACSRGLMSRAD
MLBr_02608|M.leprae_Br4923 RLDLASVFLYAGSILPGRTKLSDGTEHEVTLIDAFEAVGACSRGLMPRAD
TH_0983|M.thermoresistible__bu RLDLAAVFLYAGSILPGIAKLSDGTEREVTIIDAFEAVGACARGLMPRED
Mflv_5171|M.gilvum_PYR-GCK RLDIPTVFVSGGPMEGGSAVLVDGTVR--TRLNLVSAIADAVDSGVSDPD
Mvan_1161|M.vanbaalenii_PYR-1 RLNIPTVFVSGGPMEGGTAVLVDGTVR--TRLNLISAIADAVDTTVSDRD
MSMEG_1290|M.smegmatis_MC2_155 RLNVPTVFVSGGPMEGGRATLVDGRVR--TGLHLIDSMSGAADPTTSDED
MAB_4640c|M.abscessus_ATCC_199 RLNIPAVFVSGGPMEGGRATLRDGTVR--TGMTLITAVAESASDSVSDED
**::.:**: .*.: * : * ** : * : . ::. . . *
MMAR_0432|M.marinum_M VDAIERAICPGEGACGGMYTANTMASAAEALGMSLPGSAAPPATDRRRDG
MUL_1082|M.ulcerans_Agy99 VDAIERAICPGEGACGGMYTANTMASAAEALGMSLPGSAAPPATDRRRDG
MAV_4989|M.avium_104 VDAIERAICPGEGACGGMYTANTMASAAEALGMSLPGSAAPPATDRRRDG
Mb0195c|M.bovis_AF2122/97 VDAIERAICPGEGACGGMYTANTMASAAEALGMSLPGSAAPPATDRRRDG
Rv0189c|M.tuberculosis_H37Rv VDAIERAICPGEGACGGMYTANTMASAAEALGMSLPGSAAPPATDRRRDG
MLBr_02608|M.leprae_Br4923 VDAIERAICPGEGACGGMYTANTMASAAEALGMSLPGSAAPPATDRRRDG
TH_0983|M.thermoresistible__bu VDAIERAICPGEGACGGMYTANTMASAAEALGMSLPGSAAPPATDRRRDG
Mflv_5171|M.gilvum_PYR-GCK LARIEESACPTCGSCSGMFTANSMNCLTEALGLALPGNGSVLATHTARRA
Mvan_1161|M.vanbaalenii_PYR-1 LARIEESACPTCGSCSGMFTANSMNCLTEALGLALPGNGSVLATHTARRA
MSMEG_1290|M.smegmatis_MC2_155 LARIEEAACPTCGSCSGMFTANSMNCLVEALGLALPGNGSLLATHTARKA
MAB_4640c|M.abscessus_ATCC_199 LARIEDQACPTCGSCSGMFTANSMNCLTEALGLAPPGNGTTLATHTARRG
: ** ** *:*.**:***:* . .****:: **..: **. * .
MMAR_0432|M.marinum_M FARQSGQAVIELLRQ-------GITARDIMTREAFENAIAVVMAFGGSTN
MUL_1082|M.ulcerans_Agy99 FARQSGQAVIELLRQ-------GITARDIMTREAFENAIAVVMAFGGSTN
MAV_4989|M.avium_104 FARRSGQAVVELLRR-------GITARDILTKEAFENAIAVVMAFGGSTN
Mb0195c|M.bovis_AF2122/97 FARRSGQAVVELLRR-------GITARDILTKEAFENAIAVVMAFGGSTN
Rv0189c|M.tuberculosis_H37Rv FARRSGQAVVELLRR-------GITARDILTKEAFENAIAVVMAFGGSTN
MLBr_02608|M.leprae_Br4923 FARRSGQAVIELLRR-------GITARDILTKEAFENAIAVVMAFGGSTN
TH_0983|M.thermoresistible__bu FARKSGQAVVELLRR-------GITARDILTKEAFENAIAVVMAFGGSTN
Mflv_5171|M.gilvum_PYR-GCK LYENAGATVMDLCRRYYDEDDTSVLPRAIANREAFDNAMAMDMAMGGSTN
Mvan_1161|M.vanbaalenii_PYR-1 LYEDAGATVMDLCRRYYDEDDASVLPRAIANRDAFDNAMAMDIAMGGSTN
MSMEG_1290|M.smegmatis_MC2_155 LYQNAGRTVMELAERYYDQGDVSALPRAVASRAAFENAMAMDIAMGGSTN
MAB_4640c|M.abscessus_ATCC_199 LYEQAGSLIVELAQRYYGSDDTTVLPREIASRRAFENAMCMDIAMGGSTN
: . :* :::* .: .* : .: **:**:.: :*:*****
MMAR_0432|M.marinum_M AVLHLLAIAHEANVELTLDDFSRIGSKVPHLADVKPFGRHVMSHVDHIGG
MUL_1082|M.ulcerans_Agy99 AVLHLLAIAHEANVELTLDDFSRIGSKVPHLADVKPFGRHVMSHVDHIGG
MAV_4989|M.avium_104 AVLHLLAIAHEADVALSLDDFSRIGSKVPHLADVKPFGRHVMTDVDHIGG
Mb0195c|M.bovis_AF2122/97 AVLHLLAIAHEANVALSLQDFSRIGSGVPHLADVKPFGRHVMSDVDHIGG
Rv0189c|M.tuberculosis_H37Rv AVLHLLAIAHEANVALSLQDFSRIGSGVPHLADVKPFGRHVMSDVDHIGG
MLBr_02608|M.leprae_Br4923 AILHLLAIAHEADVTLSLEDFSRIGFKVPHIADVKPFGRHVMFDVDHIGG
TH_0983|M.thermoresistible__bu AVLHLLAIAHEANVKLTLEDFTRIGNKVPHLADVKPFGAHVMFDVDHIGG
Mflv_5171|M.gilvum_PYR-GCK TILHLLAAAREAELDYTLEDIEKRSRQIPCLCKVAPNGHYLMEDVHRAGG
Mvan_1161|M.vanbaalenii_PYR-1 TILHLLAAAREAELDYTLEDIEKRSRAVPCLCKVAPNGHYLMEDVHRAGG
MSMEG_1290|M.smegmatis_MC2_155 TVLHLLAAAHEADLDFAITEIDEISRRIPCLCKVAPNGSYLMEDVHRAGG
MAB_4640c|M.abscessus_ATCC_199 TVLHLLAAAHEAELDFALADIDALSRRVPCLSKVAPNGAYLVEDVHRAGG
::***** *:**:: :: :: . :* :..* * * ::: .*.: **
MMAR_0432|M.marinum_M VPVVMKTLLDAGLLHGDCLTVTG-----------------RTVAENLEAI
MUL_1082|M.ulcerans_Agy99 VPVVMKTLLDAGLLHGDCLTVTG-----------------RTVAENLEAI
MAV_4989|M.avium_104 VPVMMKALLDAGLLNGDCLTVTG-----------------ATVAQNLAAI
Mb0195c|M.bovis_AF2122/97 VPVVMKALLDAGLLHGDCLTVTG-----------------HTMAENLAAI
Rv0189c|M.tuberculosis_H37Rv VPVVMKALLDAGLLHGDCLTVTG-----------------HTMAENLAAI
MLBr_02608|M.leprae_Br4923 VPVVMKALLDAGLLHGDCLTVTG-----------------QTMAENLASI
TH_0983|M.thermoresistible__bu VPVIMKTLLDAGLLHGDCLTVTG-----------------KTMAENLAHI
Mflv_5171|M.gilvum_PYR-GCK IPAILGELWRGGHLHETVHSVHAGSLPEWLRRWDVRGGQACEEAIELFHA
Mvan_1161|M.vanbaalenii_PYR-1 IPAILGELWRGGHLHETVHSVHSRSLAQWLRRWDIRGGSASPEAVELFHA
MSMEG_1290|M.smegmatis_MC2_155 IPAILGELHRAGLLAKDVHAIHAATLDDWLDAWDIRAADPSDAAVDLFHA
MAB_4640c|M.abscessus_ATCC_199 VPALLGELNRAGLLHRDVHSVHAPDLTTWLTQWDIRGTAPSPEAIELFHA
:*.:: * .* * :: . * :*
MMAR_0432|M.marinum_M AP-------------------PDPDGKVLRALNNPIHATGGITILRGSLA
MUL_1082|M.ulcerans_Agy99 AP-------------------PDPDGKVLRALINPIHATGGITILRGSLA
MAV_4989|M.avium_104 AP-------------------PDPDGKVLRALSDPLHPTGGITILRGSLA
Mb0195c|M.bovis_AF2122/97 TP-------------------PDPDGKVLRALANPIHPSGGITILHGSLA
Rv0189c|M.tuberculosis_H37Rv TP-------------------PDPDGKVLRALANPIHPSGGITILHGSLA
MLBr_02608|M.leprae_Br4923 AP-------------------PDPDGQVIRTLHNPIHPTGGITILRGSLA
TH_0983|M.thermoresistible__bu AP-------------------PDPDGKVLRALSNPIHPTGGITILHGSLA
Mflv_5171|M.gilvum_PYR-GCK APGCVRSASAFSQSERWESLDTDASTGCIRDVRHAYSEDGGLAILRGNLS
Mvan_1161|M.vanbaalenii_PYR-1 APGCVRSASAFSQSQRWDTLDTDAEEGCIRDVQHAYSADGGLAVLRGNLA
MSMEG_1290|M.smegmatis_MC2_155 APGGRRSATAFSQSERWESLDLDAMAGCVRDVDHAYSPDGGLAILTGNLA
MAB_4640c|M.abscessus_ATCC_199 APGGERSARAFSQSQRWHTLDLDAENGCIRDTEHAYSADGGLAVLTGNLA
:* *. :* .. **:::* *.*:
MMAR_0432|M.marinum_M PEGAVVKTAGLDT--DVFEGTARVFDGERAALDALEDGTITHGDVVVIRY
MUL_1082|M.ulcerans_Agy99 PEGAVVKTAGLDT--DVFEGTARVFDGERAALDALEDGTITHGDVVVIRY
MAV_4989|M.avium_104 PEGAVVKSAGFDS--DVFEGTARVFDGERAALDALEDGTITKGDAVVIRY
Mb0195c|M.bovis_AF2122/97 PEGAVVKTAGFDS--DVFEGTARVFDGERAALDALEDGTITVGDAVVIRY
Rv0189c|M.tuberculosis_H37Rv PEGAVVKTAGFDS--DVFEGTARVFDGERAALDALEDGTITVGDAVVIRY
MLBr_02608|M.leprae_Br4923 PDGAVVKTAGLDS--DVFEGTARVFDGERAALDALKDGTIAKGDVVVIRY
TH_0983|M.thermoresistible__bu PEGAVVKSAGFDT--SVFEGTARVFDRERAAMDALEDGTIQAGDVVVIRY
Mflv_5171|M.gilvum_PYR-GCK RDGCIVKTAGVDESIWTFSGPAVVVESQEDAVDAILNGRVQPGDVVVVRF
Mvan_1161|M.vanbaalenii_PYR-1 VDGCIVKTAGVDESMLTFTGPAVVVESQEGAVEAILNGRARPGDVVVVRY
MSMEG_1290|M.smegmatis_MC2_155 PDGAVVKTAGVDENLLTFTGPAVVVESQEEAVDAILGDRITAGDVLVVRY
MAB_4640c|M.abscessus_ATCC_199 PNGCIVKTAGVDEKIRVFSGPAVVLESQEAAVEAILNDRVRPGDVVVIRY
:*.:**:**.* .* *.* *.: :. *::*: .. **.:*:*:
MMAR_0432|M.marinum_M EGPKGGPGMREMLAITGAIKGAGLGKDVLLLTDGRFSGGTTGFCVGHIAP
MUL_1082|M.ulcerans_Agy99 EGPKGGPGMREMLAITGAIKGAGLGKDVLLLTDGRFSGGTTGFCVGHIAP
MAV_4989|M.avium_104 EGPKGGPGMREMLAITGAIKGAGLGKDVLLLTDGRFSGGTTGLCVGHIAP
Mb0195c|M.bovis_AF2122/97 EGPKGGPGMREMLAITGAIKGAGLGKDVLLLTDGRFSGGTTGLCVGHIAP
Rv0189c|M.tuberculosis_H37Rv EGPKGGPGMREMLAITGAIKGAGLGKDVLLLTDGRFSGGTTGLCVGHIAP
MLBr_02608|M.leprae_Br4923 EGPKGGPGMREMLAITGAIKGAGLGKDVLLLTDGRFSGGTTGFCVGHIAP
TH_0983|M.thermoresistible__bu EGPKGGPGMREMLAITGAIKGAGLGKDVLLLTDGRFSGGTTGLCVGHVAP
Mflv_5171|M.gilvum_PYR-GCK EGPKGGPGMQEMLYPTSYLKGRGLGKVCALVTDGRFSGGSSGLAVGHVSP
Mvan_1161|M.vanbaalenii_PYR-1 EGPKGGPGMQEMLYPTSYLKGRGLATVCALVTDGRFSGGSSGLSIGHVSP
MSMEG_1290|M.smegmatis_MC2_155 EGPRGGPGMQEMLYPTSFLKGRGLGKSCALITDGRFSGGSSGLSIGHVSP
MAB_4640c|M.abscessus_ATCC_199 EGPRGGPGMQEMLYPTAFLKGRGLGASCALITDGRFSGGTSGLSIGHISP
***:*****:*** *. :** **. *:********::*:.:**::*
MMAR_0432|M.marinum_M EAVDAGPIAFVRDGDRIRLDVAGRTLDVLTDPAEFAARQQ------NFTP
MUL_1082|M.ulcerans_Agy99 EAVDAGPIAFVRDGDRIRLDVAGRTLDVLTDPPEFAARQQ------NFTP
MAV_4989|M.avium_104 EAVDAGPIAFLRDGDRIRLDVANRVLDVLVDPAEFDSRRT------AFTP
Mb0195c|M.bovis_AF2122/97 EAVDGGPIALLRNGDRIRLDVAGRVLDVLADPAEFASRQQ------DFSP
Rv0189c|M.tuberculosis_H37Rv EAVDGGPIALLRNGDRIRLDVAGRVLDVLADPAEFASRQQ------DFSP
MLBr_02608|M.leprae_Br4923 EAVEAGPIAFLRDGDRVLLDVVGCSLDVLVDPVEFSSRKK------DFIP
TH_0983|M.thermoresistible__bu EAVDGGPIAFLRDGDRIRLDVAKGTLDVLVDPAEFEARKK------DWEP
Mflv_5171|M.gilvum_PYR-GCK EAAAGGTIALVEDGDRITIDIPRRTIGLDVADDELDRRRAALEG-GGYRP
Mvan_1161|M.vanbaalenii_PYR-1 EAAAGGVIALIEDGDTITIDIPHRTIALDVGDDELQRRRAALQASGGYRP
MSMEG_1290|M.smegmatis_MC2_155 EAAAGGPIALIRDGDRVTIDIPSRTVTLDVDGDELARRRDELLASGGYRP
MAB_4640c|M.abscessus_ATCC_199 EAAAGGPIALVEDGDIINIDIARRSIVVAVDPQTLQRRRALLESGNGYRP
**. .* **::.:** : :*: : : . : *: : *
MMAR_0432|M.marinum_M P-APRYKTGVLAKYVKLVGSAAIGAVCG--------
MUL_1082|M.ulcerans_Agy99 P-APRYKTGVLAKYVKLVGSAAIGAVCG--------
MAV_4989|M.avium_104 P-PPRYKTGVLAKYVKLVGSAAIGAVCG--------
Mb0195c|M.bovis_AF2122/97 P-PPRYTTGVLSKYVKLVSSAAVGAVCG--------
Rv0189c|M.tuberculosis_H37Rv P-PPRYTTGVLSKYVKLVSSAAVGAVCG--------
MLBr_02608|M.leprae_Br4923 P-APRYTTGVLAKYVKLVSSATVGAVCG--------
TH_0983|M.thermoresistible__bu L-PPRYTTGVLAKYTKLVGSAATGAVCT--------
Mflv_5171|M.gilvum_PYR-GCK RHRERPVSAALRAYAALALSADKGAVRDVGRS----
Mvan_1161|M.vanbaalenii_PYR-1 RDRQRVVSAALRAYAALAMSADKGAVRDVSAV----
MSMEG_1290|M.smegmatis_MC2_155 RDRVRPVSAALRAYALLATSADRGAVRDVDNVVSDR
MAB_4640c|M.abscessus_ATCC_199 VTRERHVSAALRAYAAMATSADKGAVRSLSG-----
* :..* *. :. ** ***