For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKFVLASYGTRGDIEPSVVVGRELQSRGHTVLMAVPPDLIEFTEAANLRTVAYGLDTKTWLDIYRNFWTF ALHRFWKAREIRSLWREMWDLSDECWAQMNATLRSVAADADVLFAGQSYQEPAANVAEFYDVPLVTLHHI PMRPNGRLVTFLPPVLARASMTAFDWFAWRLNKKVEDAQRRDLGLPKATGPSTQRIADRMSLEVQAYDEV CFPGLAEEWAKWGELRPYVGTLTMELTTPADEEVSSWICGGTPPICFGFGSMPVESPADTVAMISSVCAE LGERALICAGWSDFSDVALADHVKVVGAVNYATVFPGCRAVVHHGGSGTTAASLRAGLPTLVLSMDANQA LWGAQIKRLKVGTTRRFSKTNRRTLIADLRTILDPAYAARAQALARGMTRPAESVVRAADVIEAFARSRC SA
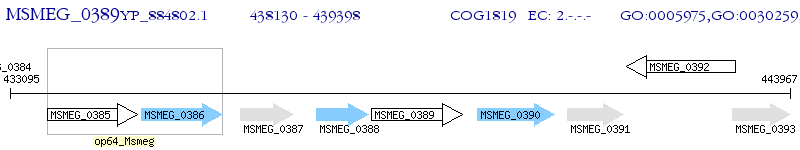
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0389 | - | - | 100% (422) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_0385 | - | e-145 | 59.17% (409) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_0392 | - | e-128 | 55.61% (419) | glycosyl transferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1551 | - | 1e-128 | 57.86% (420) | glycosyltransferase |
| M. gilvum PYR-GCK | Mflv_0466 | - | 3e-05 | 23.75% (240) | glycosyl transferase family protein |
| M. tuberculosis H37Rv | Rv1524 | - | 1e-128 | 57.86% (420) | glycosyltransferase |
| M. leprae Br4923 | MLBr_02348 | - | 1e-124 | 54.03% (422) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_4107c | - | 0.0 | 76.30% (422) | glycosyltransferase GtfA |
| M. marinum M | MMAR_2353 | - | 1e-123 | 52.78% (413) | UDP-glycosyltransferase |
| M. avium 104 | MAV_3265 | - | 0.0 | 75.18% (419) | glycosyl transferase |
| M. thermoresistible (build 8) | TH_1989 | - | 3e-48 | 67.38% (141) | Glycosyltransferase family protein 28 |
| M. ulcerans Agy99 | MUL_1529 | - | 1e-101 | 50.97% (361) | UDP-glycosyltransferase |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1551|M.bovis_AF2122/97 MKFVVASYG-TRGDIEPCAAVGLELQRRGHDVCLAVPPNLIGFVETAGLS
Rv1524|M.tuberculosis_H37Rv MKFVVASYG-TRGDIEPCAAVGLELQRRGHDVCLAVPPNLIGFVETAGLS
MMAR_2353|M.marinum_M MKFALAING-TRGDVEPCAALGRELSRRGHEVRVAVPPNLTGLAAAAGLE
MUL_1529|M.ulcerans_Agy99 ----MSNRA-LHSGGNYRAEATKDAS--------QYPPNLTGVAAAAGLE
MLBr_02348|M.leprae_Br4923 MKFTLAASG-SRGDVEPFAALGLELQRRGHEVRIGVPPDMLRFVESAGLA
MSMEG_0389|M.smegmatis_MC2_155 MKFVLASYG-TRGDIEPSVVVGRELQSRGHTVLMAVPPDLIEFTEAANLR
MAB_4107c|M.abscessus_ATCC_199 MKFVLASYG-TRGDIEPSVVVARELQRRGHDVVMAVPPDLISFTESAGLE
MAV_3265|M.avium_104 MKCVLASYG-TRGDIEPSVVVARELQRRGHDMIMAVPPDSVSFTEAAGLT
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_0466|M.gilvum_PYR-GCK MAVILAYTAPALGHLFPFCAVLQELARRGHDIHVRTLAAGLQICRDEGFT
Mb1551|M.bovis_AF2122/97 AVAYGSRDSQEQLDEQ------FLHNAWKLQNPIKLLREAMAPVTEGWAE
Rv1524|M.tuberculosis_H37Rv AVAYGSRDSQEQLDEQ------FLHNAWKLQNPIKLLREAMAPVTEGWAE
MMAR_2353|M.marinum_M AVPYGP-DTDELLNDE-----DNLQEFWKPQTSIKLVREGISDLRQAWSD
MUL_1529|M.ulcerans_Agy99 AVPYCP-DTDELLNDE-----DNLQEFWKPQTSIKLVREGISDLRQAWSD
MLBr_02348|M.leprae_Br4923 AVAYGP-DTQEFLARDT---YSQWRQWWKILPPIKALQQ----LRQAWAD
MSMEG_0389|M.smegmatis_MC2_155 TVAYGL-DTKTWLDIYRNFWTFALHRFWKAREIRSLWREMWDLSDECWAQ
MAB_4107c|M.abscessus_ATCC_199 TVSYGL-DTKTWLDVYRNFWTFFFHTFWKVREIRTMWRQMWELSDECWAQ
MAV_3265|M.avium_104 AVPYGL-DSQAWLDVYRNFWTSFFRGFWKMGEMRGMWREMWDLSDQCWAQ
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_0466|M.gilvum_PYR-GCK AAAVDPRIEAIQADDT------------LTGTVRAAADTVEVLTARAPLE
Mb1551|M.bovis_AF2122/97 LSAMLTPVAAGADLLLTGQIYQEVVANVAEHHGIPLAALH-FYPVRANGE
Rv1524|M.tuberculosis_H37Rv LSAMLTPVAAGADLLLTGQIYQEVVANVAEHHGIPLAALH-FYPVRANGE
MMAR_2353|M.marinum_M MAGTLKTLADGADLVLTGMIHQGIALNAAEYYGIPLATMH-FLPSRVNSR
MUL_1529|M.ulcerans_Agy99 MAGTLKTLADGADLVLTGMIHQGIALNAAEYYGIPLATMH-FLPSRVNSR
MLBr_02348|M.leprae_Br4923 MATDLKSLADGADLVMTGVVYQGVVANVAEYYGIPFGVLH-FVPARVNGK
MSMEG_0389|M.smegmatis_MC2_155 MNATLRSVAADADVLFAGQSYQEPAANVAEFYDVPLVTLH-HIPMRPNGR
MAB_4107c|M.abscessus_ATCC_199 MNTTLMSAAQGADVLFAGQSYQEPAANVAEYHDIPLVTLH-HIPMRPNGQ
MAV_3265|M.avium_104 MNTTLLSLADGADLLFAGQSYQEPAANVAEYYDLPLATLH-HVPMRPNGQ
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_0466|M.gilvum_PYR-GCK VADLLSAVESVQPDVVIVDANCWGAMSVAETLGRPWVVLSPFTPYLRSPG
Mb1551|M.bovis_AF2122/97 IAFPARLPAPLVRSTITAIDWLYWRMTKGVED--AQRRELGLPKASTPAP
Rv1524|M.tuberculosis_H37Rv IAFPARLPAPLVRSTITAIDWLYWRMTKGVED--AQRRELGLPKASTPAP
MMAR_2353|M.marinum_M VIP--ILSPRLNRSVISALWWVHWRVTKNPEE--AQRRELGMAKASSSST
MUL_1529|M.ulcerans_Agy99 VIP--ILSPRLNRSVISALWWVHWRVTKNPEE--AQRRELGMAKASSSST
MLBr_02348|M.leprae_Br4923 IIP--SLPSPLNRAILATVWRAHWLLAKKPED--AQRRELGLPKATSLST
MSMEG_0389|M.smegmatis_MC2_155 LVT--FLPPVLARASMTAFDWFAWRLNKKVED--AQRRDLGLPKATGPST
MAB_4107c|M.abscessus_ATCC_199 LVT--ILPSRLGRAAMRAFDWVSWRLNKKVED--AQRRELRLPKASSPSP
MAV_3265|M.avium_104 VVS--VLPAPLARSAMTAFDWFCWRLNKKVED--AQRRELGLPKATGPSP
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_0466|M.gilvum_PYR-GCK APPFGPGATPWPGPLGRIRDWGIGLVTTAVFDRPFARQIAPLRRALGLTP
Mb1551|M.bovis_AF2122/97 RRMAVRGSLEIQAYDALCFPGLAAEW---GGRRPFVGALTMESATDADDE
Rv1524|M.tuberculosis_H37Rv RRMAVRGSLEIQAYDALCFPGLAAEW---GGRRPFVGALTMESATDADDE
MMAR_2353|M.marinum_M SRILKGRSLEIQAYDELCFPGLAAEWAEWGDRRPFVGALTLEMPTESDNE
MUL_1529|M.ulcerans_Agy99 SRILKGRSLEIQAYDELCFPGLAAEWAEWGDRRPFVGALTLEMPTESDNE
MLBr_02348|M.leprae_Br4923 RRIVKRGALEIQAYDELCFPGLAAEWAEYGDRRPFVGALTLELPTAADNE
MSMEG_0389|M.smegmatis_MC2_155 QRIADRMSLEVQAYDEVCFPGLAEEWAKWGELRPYVGTLTMELTTPADEE
MAB_4107c|M.abscessus_ATCC_199 QRIAERRSLEIQGYDSVCFPGLATEWAKWNGLRPFVGSLTMAMTQEADEE
MAV_3265|M.avium_104 RRIAERGSLEIQAYDEVCFPGLAEEWAKWDGQRPFVGTLTMELTTEADDE
TH_1989|M.thermoresistible__bu --------------------------------------------------
Mflv_0466|M.gilvum_PYR-GCK VRTAGDLLLRAPAVLVATGKPLEYPHTDWGETVTMIGPAVFEPRAAQRP-
Mb1551|M.bovis_AF2122/97 VASWIAADTPPIYFGFGSMPIGSLADRVAMISAACAELGERALICSGPSD
Rv1524|M.tuberculosis_H37Rv VASWIAADTPPIYFGFGSMPIGSLADRVAMISAACAELGERALICSGPSD
MMAR_2353|M.marinum_M VLSWIASGTPPIYFGFGSMPVESPAATIAMISAACAELGERALICTGSND
MUL_1529|M.ulcerans_Agy99 VLSWIASGTPPIYFGFGSMPVESPAATIAIISAACAELGERALICTGSND
MLBr_02348|M.leprae_Br4923 VLSWIAAGTPPIYFGFGSMPIVSPTDTVAMIAAACADLGERALISVKPKD
MSMEG_0389|M.smegmatis_MC2_155 VSSWICGGTPPICFGFGSMPVESPADTVAMISSVCAELGERALICAGWSD
MAB_4107c|M.abscessus_ATCC_199 VASWIAGGTPPIFFGFGSMPVESPSAALEMIGSACAELGERALIGAGWSD
MAV_3265|M.avium_104 VMSWIRGGTPPICFGFGSMPVESPADTVDMIGTACAQLGERALLCAGRSD
TH_1989|M.thermoresistible__bu -----------------------------MFASVCSRLGERALICAGGTD
Mflv_0466|M.gilvum_PYR-GCK --SWLDHIDDPIVLVTTSSVGQSDAALVETAVEALRDEPVHLVVTAPTGD
. : :: *
Mb1551|M.bovis_AF2122/97 ATGIPQFDHVKVVRVVSHAAVFPTCRAVVHHGGAGTTAAGLRAGIPTLIL
Rv1524|M.tuberculosis_H37Rv ATGIPQFDHVKVVRVVSHAAVFPTCRAVVHHGGAGTTAAGLRAGIPTLIL
MMAR_2353|M.marinum_M LGGLQAPENVKLVSGVSHAAVFPACRAVVHHGGAGTTAAGMRAGIPTLIL
MUL_1529|M.ulcerans_Agy99 LGGLQAPENVKLVSGVSHAAVFPACCAVVHHGGAGTTAAGMRAGIPTLIL
MLBr_02348|M.leprae_Br4923 LTQVPKFDHVKIVTSVSHAAVFPACRAVVHHGGAGTTAASLRAGVPTLIL
MSMEG_0389|M.smegmatis_MC2_155 FSDVALADHVKVVGAVNYATVFPGCRAVVHHGGSGTTAASLRAGLPTLVL
MAB_4107c|M.abscessus_ATCC_199 FGGAQLPEHVKVVGAVNYATVFPACRAVVHHGGSGTTAASLRAGVPTLIL
MAV_3265|M.avium_104 FSGVPAPDHVKVVGPVNYATIFPACRAVVHHGGSGTTAASLRAGVPTLIL
TH_1989|M.thermoresistible__bu IDGVPEYGHVKVVGAVNYARVFPACRAIVHHGGSGTTAAVLRAGVPSLVT
Mflv_0466|M.gilvum_PYR-GCK VDVMTRPG-VTQVRFAPHSMLLEHAVCVITHGGMGVTQKALARGVPVCVV
*. * . :: :: . .:: *** *.* : *:* :
Mb1551|M.bovis_AF2122/97 WVTSDQPIWAAQIKQLKVGRGRRFSSATKESLIADLRTILAPDYVTRARE
Rv1524|M.tuberculosis_H37Rv WVTSDQPIWAAQIKQLKVGRGRRFSSATKESLIADLRTILAPDYVTRARE
MMAR_2353|M.marinum_M WIWIEQPIWLAQIKRLKIGSGRRLASTTKETLVSDLRCILGPDYVARART
MUL_1529|M.ulcerans_Agy99 WIWIEQPIWLAQIKRLKIGSGRRLASTTKETLVSDLRCILGPDYVARP--
MLBr_02348|M.leprae_Br4923 WIFIEQPVWAAQIKRLKVGAGRRFSATTQRSLAADLRTILAPQYATRARE
MSMEG_0389|M.smegmatis_MC2_155 SMDANQALWGAQIKRLKVGTTRRFSKTNRRTLIADLRTILDPAYAARAQA
MAB_4107c|M.abscessus_ATCC_199 SMDANQTLWGGQVKKLKVGTTRRFSTTTRESLVADLRRILASDYADRARE
MAV_3265|M.avium_104 SMDANQTIWGAQLKRLKVGATRRFSATDRDSLVQDLRKILAPDYARRARE
TH_1989|M.thermoresistible__bu WSSADQPYWGNQVKRLKVGTSRRFSHTTPDTLVADLRRILTPDYAVRARA
Mflv_0466|M.gilvum_PYR-GCK PFGRDQFEVARRVELAGCGTRLPAGKLSVGRLRTAVKTAAT--MTGGAAR
:* ::: * . * :: . .
Mb1551|M.bovis_AF2122/97 IASRMTKPAASVTATADLLEDAARRAR--------
Rv1524|M.tuberculosis_H37Rv IASRMTKPAASVTATADLLEDAARRAR--------
MMAR_2353|M.marinum_M IANEMSKPGESATRAADLLENEVSVS---------
MUL_1529|M.ulcerans_Agy99 -------PG--LRRC--------------------
MLBr_02348|M.leprae_Br4923 VANRMSKPDESVNAAADLLEDKASRNKSRTRYQDC
MSMEG_0389|M.smegmatis_MC2_155 LARGMTRPAESVVRAADVIEAFARSRCSA------
MAB_4107c|M.abscessus_ATCC_199 LAAGMTKPGESAVAAADAMERFAQARCSA------
MAV_3265|M.avium_104 IAARMTKPADSVMKAADVVEDFVQSRSRTGRS---
TH_1989|M.thermoresistible__bu VAARMTDPADSVVRAADLFETAVECAPGVAHRE--
Mflv_0466|M.gilvum_PYR-GCK VAAGFAATG-GVARGADVVESRLVARCALPEEQ--
.