For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDTRHAPPPAPRPIPEYDVSAGPFSHLAAERDRLAALRWRHFDAEQVGTTLGAQISGVDLTRPLAGAVF DELRTALHEYKVLFFRDQPMSADAHAALARQFGPLEVHPLLPANSAESALARFQKDANVSGYENAWHHDV TWRPEPSILAILHAIEVPPIGGDTLFADMYAAYDSLDDETKAEIDRLDAAHDFTYYLRGLVPDEKIAELQ AAHPPVTHPVVCTHPITGRRHLYVNRMFVSHIAGYPHDTGRALLDRLCRTADAPEHQVRFRWAPDSVAIW DNRAVQHYAASGYWPHTRIMERASVTGPVPCR
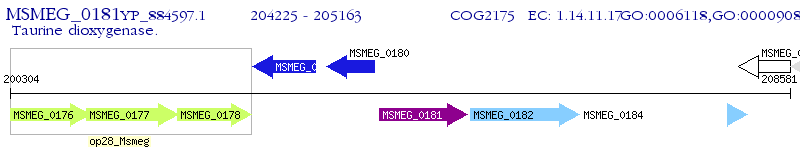
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0181 | - | - | 100% (312) | alpha-ketoglutarate-dependent taurine dioxygenase |
| M. smegmatis MC2 155 | MSMEG_3870 | - | 2e-28 | 33.57% (277) | alpha-ketoglutarate-dependent taurine dioxygenase |
| M. smegmatis MC2 155 | MSMEG_0291 | - | 5e-24 | 32.50% (280) | dioxygenase, TauD/TfdA family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3440 | - | 2e-33 | 35.40% (274) | dioxygenase |
| M. gilvum PYR-GCK | Mflv_3107 | - | 8e-33 | 32.97% (279) | taurine dioxygenase |
| M. tuberculosis H37Rv | Rv3406 | - | 2e-33 | 35.40% (274) | dioxygenase |
| M. leprae Br4923 | MLBr_01992 | - | 1e-11 | 23.84% (281) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_3621c | - | 2e-54 | 41.97% (274) | taurine dioxygenase |
| M. marinum M | MMAR_1141 | tauD | 7e-33 | 35.64% (275) | taurine catabolism dioxygenase, TauD |
| M. avium 104 | MAV_4353 | - | 9e-33 | 35.93% (270) | alpha-ketoglutarate-dependent taurine dioxygenase |
| M. thermoresistible (build 8) | TH_0024 | - | 8e-22 | 28.88% (277) | dioxygenase, TauD/TfdA family protein |
| M. ulcerans Agy99 | MUL_0904 | tauD | 5e-33 | 35.64% (275) | taurine catabolism dioxygenase, TauD |
| M. vanbaalenii PYR-1 | Mvan_4182 | - | 3e-21 | 30.47% (279) | taurine catabolism dioxygenase TauD/TfdA |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0181|M.smegmatis_MC2_155 MTDTRHAPPPAPRPIPEYDVSAGPFSHLAAERDRLAALRWRHFDAEQVGT
MAB_3621c|M.abscessus_ATCC_199 MNG------------------VAPLSKIRVQP---------------LTC
Mb3440|M.bovis_AF2122/97 -----------------------MTDLITVKK---------------LGS
Rv3406|M.tuberculosis_H37Rv -----------------------MTDLITVKK---------------LGS
MMAR_1141|M.marinum_M -----------------------MTDLITVKK---------------LGS
MUL_0904|M.ulcerans_Agy99 -----------------------MTDLITVKK---------------LGS
MAV_4353|M.avium_104 -----------------------MTGQITVTK---------------LGS
Mflv_3107|M.gilvum_PYR-GCK ------------------------MSSPDVVK---------------LGA
TH_0024|M.thermoresistible__bu ------------------------MSRLTINR---------------LTA
Mvan_4182|M.vanbaalenii_PYR-1 ------------------------MTLLTINK---------------LTS
MLBr_01992|M.leprae_Br4923 -------------------------MTLDVKG-----------------D
MSMEG_0181|M.smegmatis_MC2_155 TLGAQISGVDLTRP-LAGAVFDELRTALHEYKVLFFR-DQPMSADAHAAL
MAB_3621c|M.abscessus_ATCC_199 TIGAELFDVDLAEASRSDELFGEIRTLLLEHKVLFLR-DQNITRGEHVAL
Mb3440|M.bovis_AF2122/97 RIGAQIDGVRLGGD-LDPAAVNEIRAALLAHKVVFFRGQHQLDDAEQLAF
Rv3406|M.tuberculosis_H37Rv RIGAQIDGVRLGGD-LDPAAVNEIRAALLAHKVVFFRGQHQLDDAEQLAF
MMAR_1141|M.marinum_M RIGAQVDGVSLGAD-LDAAAVDQIRAALLEHKVIFFRNQHHLDDQQQLQF
MUL_0904|M.ulcerans_Agy99 RIGAQVDGVSLGAD-LDAAAVDQIRAALLEHKVIFFRNQHHLDDQQQLQF
MAV_4353|M.avium_104 RIGARVDGVRLGGD-LDDATVEQIRRALLTHKVIFFRHQHHLDDSRQLEF
Mflv_3107|M.gilvum_PYR-GCK AIGARIDGVDLARG-IDAGTAAQINAALLEHKVIFFRGQHDLDDDGQLEF
TH_0024|M.thermoresistible__bu SVGAEVTGTTADELAADAALGAAVLDALEEHGVLVFR-GLRIDPQTQVKF
Mvan_4182|M.vanbaalenii_PYR-1 SVGAEVTGLDPGRLAGDDALGTAVLDALEDNGVLVFP-GLHLEPEVQVEF
MLBr_01992|M.leprae_Br4923 GLGAQVTGVD--PKNLDSISTAEIRKIVYVNKLVVLK-DVHPTPEEFIKL
:**.: . : : ::.: :
MSMEG_0181|M.smegmatis_MC2_155 ARQFGPL--EVHPLLPANSAESALARFQKDANVS--GYENAWHHD---VT
MAB_3621c|M.abscessus_ATCC_199 ARRFGPL--EDHPVAGSDPDHPGLVRIYKEIDSAPEHYENSYHCD---AT
Mb3440|M.bovis_AF2122/97 AGLLGTP--IGHPAAIALAD-DAPIITPINSEFG---KANRWHTD---VT
Rv3406|M.tuberculosis_H37Rv AGLLGTP--IGHPAAIALAD-DAPIITPINSEFG---KANRWHTD---VT
MMAR_1141|M.marinum_M AGLLGTP--IGHPAAAAALP-DAPIITPINSEWG---KANRWHTD---VT
MUL_0904|M.ulcerans_Agy99 AGLLGTP--IGHPAAAAALP-DAPIITPINSEWG---KANRWHTD---VT
MAV_4353|M.avium_104 ARLLGTP--IGHPAASALAAKHLPVITPIDSEYG---KATRWHTD---VT
Mflv_3107|M.gilvum_PYR-GCK ARALGTPT-TAHPTVTSRGA----KVLPIDSRYD---KADSWHTD---VT
TH_0024|M.thermoresistible__bu CRHLGEVDHSSDGHHPVAGIYPVTLDKSKNSSAAYLRATFDWHIDGCTPT
Mvan_4182|M.vanbaalenii_PYR-1 CRRLGEIDFSSDGHHRVAGIYPVTLDKSKNSSAAYLRATFDWHIDGCTPT
MLBr_01992|M.leprae_Br4923 GRIIGEIVPYYEPIYR-HKDYPEIFVSSTEEGQGVPNTGAFWHVD---YI
:* . : :* *
MSMEG_0181|M.smegmatis_MC2_155 WRPEPSILAILHAIEVPPIGGDTLFADMYAAYDSLDDETKAEIDRLDAAH
MAB_3621c|M.abscessus_ATCC_199 WRESPPMGAVLRCVETPPVGGDTIWVNMVEAYRRLPAAVRDRIRDLRARH
Mb3440|M.bovis_AF2122/97 FAANYPAASVLRAVSLPSYGGSTLWANTAAAYAELPEPLKCLTENLWALH
Rv3406|M.tuberculosis_H37Rv FAANYPAASVLRAVSLPSYGGSTLWANTAAAYAELPEPLKCLTENLWALH
MMAR_1141|M.marinum_M FAANYPAASILRAVTLPNYGGSTLWANTATAYAELPEPLKCLVENLWALH
MUL_0904|M.ulcerans_Agy99 FAANYPAASILRAVTLPNYGGSTLWANTATAYAELPEPLKCLVENLWALH
MAV_4353|M.avium_104 FAANYPAASILRAVTLPSYGGSTLWASTVAAYQQLPEPLRHLTENLWALH
Mflv_3107|M.gilvum_PYR-GCK FVDRIPKASLLRAVTLPAYGGTTAWASTEAAYDRLPAPLRALTENLWAVH
TH_0024|M.thermoresistible__bu GDEYPQKATVLSAVQVADRGGETEFASSYGAYEALSEEEKRHVETLRVVH
Mvan_4182|M.vanbaalenii_PYR-1 GDDYPQMATVLSAKQVAESGGETEFASSYAAYDALTDAEKQRFASLRVVH
MLBr_01992|M.leprae_Br4923 FMPEPFAFSMTLPLAMPGHDRGTHFIDLSQVWESLPNAMKDSVRGTYSNH
:: . . * : . .: * : *
MSMEG_0181|M.smegmatis_MC2_155 DFTYYLRG----------LVPDEKIAELQAAHPPVTHPVVCTHPITGRRH
MAB_3621c|M.abscessus_ATCC_199 SIEASFGA----------AMSMRQRHELHERYPDAEHPVVRTHPETGEEI
Mb3440|M.bovis_AF2122/97 TNRYDYVT-----TKPLTAAQRAFRQVFEKPDFRTEHPVVRVHPETGERT
Rv3406|M.tuberculosis_H37Rv TNRYDYVT-----TKPLTAAQRAFRQVFEKPDFRTEHPVVRVHPETGERT
MMAR_1141|M.marinum_M TNRYDYVANE--AVQALTDTQQAFRQAFQKPDFRTEHPVVRVHPETGERT
MUL_0904|M.ulcerans_Agy99 TNRYDYVANE--AVQALTDTQQAFRQAFQKPDFRTEHPVVRVHPETGERT
MAV_4353|M.avium_104 TNRYDYVR-----NDPMNDTQRAFRQAFEKPDFRTEHPVVRVHPETGERA
Mflv_3107|M.gilvum_PYR-GCK TNTYDYAADADARLVPLADTERQYREEFVSDYYETEHPVVRVHPETGRKV
TH_0024|M.thermoresistible__bu SLEASQRR--------VTPDPTPEQLARWRARPTHEHPLVWTH-RNGRKS
Mvan_4182|M.vanbaalenii_PYR-1 SLEASQRR--------VNPDPSPEELARWRARPTHEHPLVWTH-RTGRKS
MLBr_01992|M.leprae_Br4923 SPRRYIKIRP----SDVYRPVGEVLAEIEEVTPPQKWPTVIKHPKTGQEI
* * * .*..
MSMEG_0181|M.smegmatis_MC2_155 LYVNRMFVSHIAGYPHDT-----------GRALLDRLCRTADAPEHQVR-
MAB_3621c|M.abscessus_ATCC_199 LFVN-SFTTHLVNYHRPENVRFGIDYAPGASELLQYLMRQATVPEFQVR-
Mb3440|M.bovis_AF2122/97 LLAG-DFVRSFVGLDSHE-----------SRVLFEVLQRRITMPENTIR-
Rv3406|M.tuberculosis_H37Rv LLAG-DFVRSFVGLDSHE-----------SRVLFEVLQRRITMPENTIR-
MMAR_1141|M.marinum_M LLAG-DFVRGFVGLDSQE-----------SSALFELLQRRITSPENTIR-
MUL_0904|M.ulcerans_Agy99 LLAG-DFVRGFVGLDSQE-----------SSALFELLQRRITSPENTIR-
MAV_4353|M.avium_104 LLAG-DFVRGFVGLDGHE-----------SSVLLELLQRRITMPENTVR-
Mflv_3107|M.gilvum_PYR-GCK LLLG-HFVKHFVGLGQAE-----------STALLQLLQNRVTKLENTIR-
TH_0024|M.thermoresistible__bu LVLG-ASADYVVGMDLDE-----------GRALLADLLDRATTEDRVYR-
Mvan_4182|M.vanbaalenii_PYR-1 LVLG-ASADYIVGMDPEE-----------GRALLSELLDRATTADKVYS-
MLBr_01992|M.leprae_Br4923 LYICEAATVSVEDKNGNLL----------DPMVLQELLTASGQLDPDCKS
* . . . :: * :
MSMEG_0181|M.smegmatis_MC2_155 -----FRWAPDSVAIWDNRAVQHYAASGYWPHTRIMERASVTGPVPCR--
MAB_3621c|M.abscessus_ATCC_199 -----WRWTPNSVAIWDNRSTQHYAVQDYWPAVRKMERAGICGDIPFS--
Mb3440|M.bovis_AF2122/97 -----WNWAPGDVAIWDNRATQHRAIDDYDDQHRLMHRVTLMGDVPVDVY
Rv3406|M.tuberculosis_H37Rv -----WNWAPGDVAIWDNRATQHRAIDDYDDQHRLMHRVTLMGDVPVDVY
MMAR_1141|M.marinum_M -----WNWESGDVAIWDNRATQHRAIDDYDDQHRLLHRVTLMGDVPVDVH
MUL_0904|M.ulcerans_Agy99 -----WNWESGDVAIWDNRATQHRAIDDYDDQHRLLHRVTLMGDVPVDVH
MAV_4353|M.avium_104 -----WSWAPGDVAMWDNRATQHRAIDDYDDQPRLMHRITLMGDVPVNVH
Mflv_3107|M.gilvum_PYR-GCK -----WSWELGDIAVWDNRATQHYAVSDYDDQFRRLSRVTLAGDIPVDVY
TH_0024|M.thermoresistible__bu -----HRWSVGDTVIWDNRGVLHRAAPYPEDSPREMLRTTVLGDEPIQ--
Mvan_4182|M.vanbaalenii_PYR-1 -----HHWAVGDTVIWDNRGVLHRAARYDENSPREMLRTTVLGDEPIQ--
MLBr_01992|M.leprae_Br4923 LLIHTQHYEVGDVVLWDNRALVHRAKHGTVSGTLITYRLTLLDGLKTPGY
: .. .:****. * * * : .
MSMEG_0181|M.smegmatis_MC2_155 ----------------------------
MAB_3621c|M.abscessus_ATCC_199 ----------------------------
Mb3440|M.bovis_AF2122/97 GQASRVISG---------APMEIAG---
Rv3406|M.tuberculosis_H37Rv GQASRVISG---------APMEIAG---
MMAR_1141|M.marinum_M GQRSRVISG---------APLALAG---
MUL_0904|M.ulcerans_Agy99 GQRSRVISG---------APLALAG---
MAV_4353|M.avium_104 GERSRVISG---------APLEVLAS--
Mflv_3107|M.gilvum_PYR-GCK GRRSRVIAGDASTYSQVVVPLKLAGLAS
TH_0024|M.thermoresistible__bu ----------------------------
Mvan_4182|M.vanbaalenii_PYR-1 ----------------------------
MLBr_01992|M.leprae_Br4923 AV--------------------------