For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NIHHRYATIDGQRIFYRAAGPADAPPIVLLHGFPTSSFMFRDLIPLLARNHRVIAPDHLGFGFSDAPSAD EFDYTFDALADLTEGLLTHLGIVRYAIYVQDYGAPIGWRLALRHPEAISAIVTQNGNGYVEGFVDGFWST VWDYHREQTPQTEAAIRTALDPESVKWQYVTGVADESLVSPDTWNHDAAQLARPGNDVIQLKLFADYATN PPLYPTLHEYLRTSGVPVLAVWGRNDPIFGPDGARAFEKDATDAEIHLLDGGHFLLETHVDEVAGLIDSF LDRKVS*
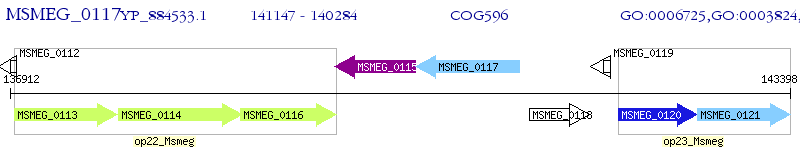
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0117 | - | - | 100% (287) | hydrolase |
| M. smegmatis MC2 155 | MSMEG_6151 | - | 6e-50 | 38.25% (285) | alpha/beta hydrolase fold-1 |
| M. smegmatis MC2 155 | MSMEG_6314 | - | 1e-18 | 30.96% (281) | haloalkane dehalogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2318 | - | 6e-21 | 44.35% (115) | haloalkane dehalogenase |
| M. gilvum PYR-GCK | Mflv_4848 | - | 1e-118 | 69.01% (284) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0134 | ephF | 3e-26 | 30.61% (294) | epoxide hydrolase EphF |
| M. leprae Br4923 | MLBr_00862 | ephD | 9e-09 | 30.36% (112) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0528c | - | 1e-14 | 34.68% (124) | epoxide hydrolase EphA |
| M. marinum M | MMAR_0342 | ephF | 1e-24 | 28.57% (287) | epoxide hydrolase EphF |
| M. avium 104 | MAV_5163 | - | 1e-21 | 29.11% (292) | hydrolase, alpha/beta fold family protein |
| M. thermoresistible (build 8) | TH_0992 | ephA | 6e-13 | 36.44% (118) | PROBABLE EPOXIDE HYDROLASE EPHA (EPOXIDE HYDRATASE) |
| M. ulcerans Agy99 | MUL_4778 | ephF | 7e-25 | 28.57% (287) | epoxide hydrolase EphF |
| M. vanbaalenii PYR-1 | Mvan_4733 | - | 3e-80 | 51.76% (284) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0342|M.marinum_M -------------MSTLPALDGVEHRFVDLDGGITIHVADAGPADGPAVM
MUL_4778|M.ulcerans_Agy99 -------------MSTLPALDGVEHRFVDLDGGITIHVADAGPADGPAVM
Rv0134|M.tuberculosis_H37Rv -------------MIALPALEGVEHRHVDVAEGVRIHVADAGPADGPAVM
MAV_5163|M.avium_104 -------------MVTMPELDGVEHRYVDLGDDVTIHVADAGPASGPAVM
MSMEG_0117|M.smegmatis_MC2_155 --------------------MNIHHRYATIDG-QRIFYRAAGPADAPPIV
Mflv_4848|M.gilvum_PYR-GCK -------------------MTVVHHRIDTVDG-HQVFYREAGDPDAPVIV
Mvan_4733|M.vanbaalenii_PYR-1 -----------------MAETCTHYRTASIDG-LDVFYREAGDRSNPTLL
MAB_0528c|M.abscessus_ATCC_199 ---------------MTDAAVDIRERDIATNG-VHLRIVEAGEPGQPAVI
TH_0992|M.thermoresistible__bu -------------------VITPSERSVETNG-VRLRLVEAGERGDPLVV
MLBr_00862|M.leprae_Br4923 -------------MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIV
Mb2318|M.bovis_AF2122/97 MDVLRTPDSRFEHLVGYPFAPHYVDVTAGDTQPLRMHYVDEGPGDGPPIV
: * * ::
MMAR_0342|M.marinum_M LVHGFPENWWEWHELIGPLAADGYRVLCPDLRGAGWSSAP---RSRYTKK
MUL_4778|M.ulcerans_Agy99 LVHGFPENWWEWHELIGPLAADGYRVLCPDLRGAGWSSAP---RSRYTKK
Rv0134|M.tuberculosis_H37Rv LVHGFPQNWWEWRDLIGPLAADGNRVLCPDLRGAGWSSAP---RSRYTKT
MAV_5163|M.avium_104 LVHGFPQNWWEWRALIGPLAADGYRVLCPDLRGAGWSSAP---RSSYRKD
MSMEG_0117|M.smegmatis_MC2_155 LLHGFPTSSFMFRDLIPLLARN-HRVIAPDHLGFGFSDAPSADEFDYTFD
Mflv_4848|M.gilvum_PYR-GCK LLHGFPTSSFMFRNLIPELADR-YRVIAPDYLGFGYSDAPTVEDFDYTFD
Mvan_4733|M.vanbaalenii_PYR-1 LLHGFPSSSHMFRNLITALADD-YHLVAPDHIGFGQSSMPSVNQFTYSFD
MAB_0528c|M.abscessus_ATCC_199 LSHGFPELAYSWRHQIPALAAAGYHVIAPDQRGYGRSSMP-AHIDDYNIE
TH_0992|M.thermoresistible__bu LAHGFPELAYSWRHQIPALVDAGYHVMAPDQRGYGGSSAP-EAIEAYDIT
MLBr_00862|M.leprae_Br4923 LVHGFPDSHVLWDGVVPLLAKR-FRIVRYDNRGVGQSSVP-KPVSAYTMT
Mb2318|M.bovis_AF2122/97 LLHGEPTWSYLYRTMIPPLSAAGHRVLAPDLIGFGRSDKP-TRIEDYTYL
* ** * : : * ::: * * * *. * *
MMAR_0342|M.marinum_M EMADDLAGVLDRLGIAS-VRLVAHDWGGPVAFIMMLRY--PAKVTGFFGM
MUL_4778|M.ulcerans_Agy99 EMADDLAGVLDRLGIAS-VRLVAHDWGGPVAFIMMLRY--PAKVTGFFGM
Rv0134|M.tuberculosis_H37Rv EMADDLAAVLDGLGVAK-VKLVAHDWGGPVAFIMMLRH--PEKVTGFFGV
MAV_5163|M.avium_104 EMADDLAGVLDRLGVRS-VDLVAHDWGGPVAFIMMLRH--PEKVTGFFGV
MSMEG_0117|M.smegmatis_MC2_155 ALADLTEGLLTHLGIVR-YAIYVQDYGAPIGWRLALRH--PEAISAIVTQ
Mflv_4848|M.gilvum_PYR-GCK ALTDSVAGLLTQLGIRR-YAIYVQDYGAPVGWRLALRD--PAAITAIITQ
Mvan_4733|M.vanbaalenii_PYR-1 RLTDITEKLIDRLGLGR-FAIYIHDYGAPIGLRIASSR--PERITGIITQ
MAB_0528c|M.abscessus_ATCC_199 ALSDDLLGILDDVGAGK-ATFVGHDWGAVVTWHTALAV--PERVSGVVGL
TH_0992|M.thermoresistible__bu RLTADLMGLLDDIGAEK-AAFIGHDWGALVVWNAALLY--PDRVAAVAGL
MLBr_00862|M.leprae_Br4923 RFADDFAAVIDELSPDRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSV
Mb2318|M.bovis_AF2122/97 RHVEWVTSWFENLDLHD-VTLFVQDWGSLIGLRIAAEHG--DRIARLVVA
: :. . :*:*. :: .
MMAR_0342|M.marinum_M NTAAPWFPR-------------------DLGMVRDIWRFWYQIPIMLPVI
MUL_4778|M.ulcerans_Agy99 NTAAPWFPR-------------------DLGMVRDIWRFWYQIPILLPVI
Rv0134|M.tuberculosis_H37Rv NTVAPWVKR-------------------DLGMLRNMWRFWYQIPMSLPVI
MAV_5163|M.avium_104 NTSAPFVKR-------------------SLSTIRNVWRFWYQIPISLPVI
MSMEG_0117|M.smegmatis_MC2_155 NGNGYVEG-------------------FVDGFWSTVWDYHREQTPQTEAA
Mflv_4848|M.gilvum_PYR-GCK NGNGYNDG-------------------FVAEGWQPAWDYQREQTPETEAA
Mvan_4733|M.vanbaalenii_PYR-1 SGNAYTEG-------------------FTP-FWDRLFAHAKDRG-ANEDA
MAB_0528c|M.abscessus_ATCC_199 SVPFTRRSQVPPTQAWNKLFG---DNFFYILYFQEPGVADADLNRDPATT
TH_0992|M.thermoresistible__bu SVPPVPRSLTRPTEAFRALVGE--DNFFYILYFQEPGVADAELDGDPART
MLBr_00862|M.leprae_Br4923 SGPSQDQLVDYILSGLRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMV
Mb2318|M.bovis_AF2122/97 NGFLPAAQG-------------------RTPLPFYVWRAFARYSPVLPAG
.
MMAR_0342|M.marinum_M GPRVIA--------------------------------DPKARYFRVLTS
MUL_4778|M.ulcerans_Agy99 GPRVIA--------------------------------DPKARYFRVLTS
Rv0134|M.tuberculosis_H37Rv GPRVIS--------------------------------DPKGRYFRLLTG
MAV_5163|M.avium_104 GPRVIS--------------------------------TPDSRFVRLLGS
MSMEG_0117|M.smegmatis_MC2_155 IRTALD--------------------------------PESVKWQYVTGV
Mflv_4848|M.gilvum_PYR-GCK LRGILT--------------------------------FETTRMQYTAGV
Mvan_4733|M.vanbaalenii_PYR-1 VRDYFT--------------------------------PETTKWQYTHGV
MAB_0528c|M.abscessus_ATCC_199 MRRMMAGMAR-------------------IDGAT-MIAPGPAGFVERMPD
TH_0992|M.thermoresistible__bu MRRMFGGLTSD------------------PDAAHRMLQPGPAGFIDRLPE
MLBr_00862|M.leprae_Br4923 LWIALSSATIRRTMVDNISTDQIHHSATLARDAAHSVKTYPANYFRAFSG
Mb2318|M.bovis_AF2122/97 RLVNFG------------------------------------TVHRVPAG
:
MMAR_0342|M.marinum_M --------WVGGGFTIADEDVRMYVERMRQPGHAVAGSRWYRTFQSSEAL
MUL_4778|M.ulcerans_Agy99 --------WVGGGFTIADEDVRMYVERMRQPGHAVAGSRWYRTFQSSEAL
Rv0134|M.tuberculosis_H37Rv --------WVGGGFRVPDDDVRLYLDCMREPGHAEAGSRWYRTFQTREML
MAV_5163|M.avium_104 --------WVGGGYTLPEEDVRLYLECMRQPGHAEAGSRWYRSFQTREML
MSMEG_0117|M.smegmatis_MC2_155 --------ADE--SLVSPDTWNHDAAQLARPGNDVIQLKLFADYATNPPL
Mflv_4848|M.gilvum_PYR-GCK --------PDP--SVVSPDTWHHDFALLSRPGNDRIQLKLFLDYSTNPKI
Mvan_4733|M.vanbaalenii_PYR-1 --------PADRLDRIAPETWLLDQAGLDRKGNDAIQLQLFWDYQFNLDG
MAB_0528c|M.abscessus_ATCC_199 --------PGELPEWLSQDELDHYITEFARTG-FTGGLNWYRNFDRNWAL
TH_0992|M.thermoresistible__bu --------PEALPDWLTAEELDHYIAEFTRTG-FTGGLNWYRNMDRNWEL
MLBr_00862|M.leprae_Br4923 RRRGKGVPVVNVPVQLIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHF
Mb2318|M.bovis_AF2122/97 ----------------VRAGYDAPFPDKTYQAGARAFPRLVPTSPDDPAV
. .
MMAR_0342|M.marinum_M PWMRGEYAD-ARVDVPVRWLHGTEDPVLTPQLLRGYEDHISD-FEVEFVD
MUL_4778|M.ulcerans_Agy99 PWMRGEYAD-ARVDVPVRWLHGTDDPVLTPQLLRGYEDHISD-FEVEFVD
Rv0134|M.tuberculosis_H37Rv RWLRGEYND-ARVDVPVRWLHGTGDPVITPDLLDGYAERASD-FEVELVD
MAV_5163|M.avium_104 SWMRGEYDG-ARVHVPVRWLSGTEDPVLTPDLLDGYAERIDD-FQLELVD
MSMEG_0117|M.smegmatis_MC2_155 YPTLHEYL--RTSGVPVLAVWGRNDPIFGPDGARAFEKDATD--AEIHLL
Mflv_4848|M.gilvum_PYR-GCK YPALHDYL--RASGVPLLAVWGDKDPFFGPDGARAFAKDAVD--SEIHLI
Mvan_4733|M.vanbaalenii_PYR-1 YPKFQEYF--RVHQPPLLVTWGRNDEIFGAAGAEAFKRDLPD--GEFHLL
MAB_0528c|M.abscessus_ATCC_199 TERLAG----ANVVVPSLFIAGAADPVLGFTDHAGSAKYRTDNRGDLLID
TH_0992|M.thermoresistible__bu TEHLAG----ATITAPALFLAGAADPVLGFMRPERATEVAVGPYRQVLLD
MLBr_00862|M.leprae_Br4923 SPMSHPQVM-AAAVHDLADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVT
Mb2318|M.bovis_AF2122/97 PANRAAWEALGRWDKPFLAIFGYRDPILGQADGPLIKHIPGAAGQPHARI
. :
MMAR_0342|M.marinum_M GVGHWIVEQRPDLVLDRLRAFLRIET------------------------
MUL_4778|M.ulcerans_Agy99 GVGHWIVEQRPDLVLDRLRAFLRIET------------------------
Rv0134|M.tuberculosis_H37Rv GVGHWIVEQRPELVLDRVRAFLAAGTEQRD--------------------
MAV_5163|M.avium_104 GVGHWIVDQRPDLVLDRVRAFLRRET------------------------
MSMEG_0117|M.smegmatis_MC2_155 DGGHFLLETHVDEVAGLIDSFLDRKVS-----------------------
Mflv_4848|M.gilvum_PYR-GCK DGGHFLLESELTRVANLIRDFLERRL------------------------
Mvan_4733|M.vanbaalenii_PYR-1 DTGHFALETHGDEIAGHIRDFLGRQEGLIRE-------------------
MAB_0528c|M.abscessus_ATCC_199 GAGHWIQQERPLEVNAALLAFLREVVH-----------------------
TH_0992|M.thermoresistible__bu GAGHWVQQERPQEVNAALIDFLRGLELQ----------------------
MLBr_00862|M.leprae_Br4923 GAGSGIGRETALALARRGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYAL
Mb2318|M.bovis_AF2122/97 KASHFIQEDSGTELAERMLSWQQAT-------------------------
. :
MMAR_0342|M.marinum_M --------------------------------------------------
MUL_4778|M.ulcerans_Agy99 --------------------------------------------------
Rv0134|M.tuberculosis_H37Rv --------------------------------------------------
MAV_5163|M.avium_104 --------------------------------------------------
MSMEG_0117|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4848|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4733|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0528c|M.abscessus_ATCC_199 --------------------------------------------------
TH_0992|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DVSDAAAVEAFAEQVSAKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLA
Mb2318|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0342|M.marinum_M --------------------------------------------------
MUL_4778|M.ulcerans_Agy99 --------------------------------------------------
Rv0134|M.tuberculosis_H37Rv --------------------------------------------------
MAV_5163|M.avium_104 --------------------------------------------------
MSMEG_0117|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4848|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4733|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0528c|M.abscessus_ATCC_199 --------------------------------------------------
TH_0992|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 VNLGGVVNCCRAFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAA
Mb2318|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0342|M.marinum_M --------------------------------------------------
MUL_4778|M.ulcerans_Agy99 --------------------------------------------------
Rv0134|M.tuberculosis_H37Rv --------------------------------------------------
MAV_5163|M.avium_104 --------------------------------------------------
MSMEG_0117|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4848|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4733|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0528c|M.abscessus_ATCC_199 --------------------------------------------------
TH_0992|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 TYMFSDCLRAELDAVGVGLTTICPGVIDTNIIQSTRIDTPTAKQERVRDR
Mb2318|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0342|M.marinum_M --------------------------------------------------
MUL_4778|M.ulcerans_Agy99 --------------------------------------------------
Rv0134|M.tuberculosis_H37Rv --------------------------------------------------
MAV_5163|M.avium_104 --------------------------------------------------
MSMEG_0117|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4848|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4733|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0528c|M.abscessus_ATCC_199 --------------------------------------------------
TH_0992|M.thermoresistible__bu --------------------------------------------------
MLBr_00862|M.leprae_Br4923 RGQLDMMFRLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLP
Mb2318|M.bovis_AF2122/97 --------------------------------------------------
MMAR_0342|M.marinum_M ------------
MUL_4778|M.ulcerans_Agy99 ------------
Rv0134|M.tuberculosis_H37Rv ------------
MAV_5163|M.avium_104 ------------
MSMEG_0117|M.smegmatis_MC2_155 ------------
Mflv_4848|M.gilvum_PYR-GCK ------------
Mvan_4733|M.vanbaalenii_PYR-1 ------------
MAB_0528c|M.abscessus_ATCC_199 ------------
TH_0992|M.thermoresistible__bu ------------
MLBr_00862|M.leprae_Br4923 QVLRSTARIRVI
Mb2318|M.bovis_AF2122/97 ------------