For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HLDELSWFVVLAETEHVTDAAAELGISQPTLSRALARLEDEVGVPLFDRVNRRLQLNAYGEILLEHARRT ITEMRSATERIAAMRDPDTGLVRLAFLHSQAGSFVPDLLRRFRAHAPHVQFSLTQGAAHDLLERLAGGQV DLAITSPRPEGFQWRGLYVERLCLAVPRGHRLASRSRITLADAGDEPFVGLAPGFGLRQLTEELCAEAGI TPHVVFEATEIPTMEGLVAAGFGVAVVPVPHAGRAEPGAEYIALAQASAKRQLGLAWPADRPLPPAAERF AQFVIRTTHDVE*
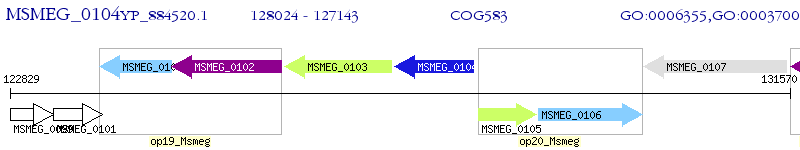
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0104 | - | - | 100% (293) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_5654 | - | 2e-35 | 34.91% (275) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_3840 | - | 7e-30 | 31.69% (284) | LysR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0121 | oxyS | 2e-15 | 26.41% (284) | oxidative stress response regulatory protein OXYS |
| M. gilvum PYR-GCK | Mflv_3565 | - | 3e-29 | 32.75% (287) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0117 | oxyS | 2e-15 | 26.41% (284) | oxidative stress response regulatory protein OXYS |
| M. leprae Br4923 | MLBr_02041 | oxyR | 7e-25 | 29.69% (293) | putative LysR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2707 | - | 8e-63 | 43.21% (287) | putative transcriptional regulator |
| M. marinum M | MMAR_4157 | - | 7e-38 | 33.80% (287) | LuxR family transcriptional regulator |
| M. avium 104 | MAV_1538 | - | 1e-67 | 46.13% (284) | transcriptional regulator, LysR family protein |
| M. thermoresistible (build 8) | TH_3504 | - | 3e-11 | 27.16% (243) | PUTATIVE putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_4020 | - | 1e-37 | 33.80% (287) | LuxR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1027 | - | 5e-29 | 33.57% (280) | LysR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2707|M.abscessus_ATCC_1997 -------MAEGDYEWYITLAELQNVTAAADQLRLAQPTLTRMLARLERRL
MAV_1538|M.avium_104 -------MAVGDYEWFITLAELQHVTAAAEQLHVAQPTLTRMLARLEQRL
MSMEG_0104|M.smegmatis_MC2_155 -------MHLDELSWFVVLAETEHVTDAAAELGISQPTLSRALARLEDEV
MMAR_4157|M.marinum_M -------MELYQLRYFQAVAECGTLRGASEQLLVSQSAVSRAITLLEAEI
MUL_4020|M.ulcerans_Agy99 -------MELYQLRYFQAVAECGTLRGASEQLLVSQSAVSRAITLLEAEI
Mb0121|M.bovis_AF2122/97 -------MLFRQLEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLEREL
Rv0117|M.tuberculosis_H37Rv -------MLFRQLEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLEREL
Mflv_3565|M.gilvum_PYR-GCK -------MELRQLEYFVAIVEEGGFTRAASRERVAQPAVSAQIRRLERSV
Mvan_1027|M.vanbaalenii_PYR-1 ------MFSLARLSCFVAVAEELHFGRAAERLHMTQPPLSRQVQQLENEL
MLBr_02041|M.leprae_Br4923 MSDKSYQPTIAGLRAFVAVVEKGHFSAAASFLGVRQSTLSQALAALESGL
TH_3504|M.thermoresistible__bu --VPSPKIDLRRLAQFVAVAELGSFTRAAATLHLSQQALSSAVRALENDV
: .:.: . *: : * .:: : ** :
MAB_2707|M.abscessus_ATCC_1997 GVQLFDRHGKRLALNPFGRIFYQHARRAQLELDSARRAIDDLANPAEGEI
MAV_1538|M.avium_104 GVALFDRHGRRLSLNTYGRIFYEHARRAQLELDSARRAIADLADPAVGEI
MSMEG_0104|M.smegmatis_MC2_155 GVPLFDRVNRRLQLNAYGEILLEHARRTITEMRSATERIAAMRDPDTGLV
MMAR_4157|M.marinum_M GVQLFTRRGRTNELNRYGQALLVGSRLAQRSVSSAIDNVRELAGVSGGTV
MUL_4020|M.ulcerans_Agy99 GVQLFTRRGRTNELNRYGQALLVGSRLAQRSVSSAIDNVRELAGVSGGTV
Mb0121|M.bovis_AF2122/97 NVTLINRGHSFEGLTREGERLVVWAKRILAEHAAFKAEVDAVRSGITGTL
Rv0117|M.tuberculosis_H37Rv NVTLINRGHSFEGLTREGERLVVWAKRILAEHAAFKAEVDAVRSGITGTL
Mflv_3565|M.gilvum_PYR-GCK GQPLLTRTSREVRLTRAGEALLPHARAALAAVRDGRAAVDDVAGLLRGSV
Mvan_1027|M.vanbaalenii_PYR-1 GVQLIDRTTRNVTLTPAGAAFLPDARRILQLAEGAAQTVKRIPAGDLGTV
MLBr_02041|M.leprae_Br4923 GVQLIERSTRRVFVTTQGRHLLPRAQAAIEAMDAFTAAAAGESDPLRGGM
TH_3504|M.thermoresistible__bu GADLFTRDGRRISLTTAGRTLLAEGKPLLAAARTVAEHVRQTEAAAQAHY
. *: * :. * : .: .
MAB_2707|M.abscessus_ATCC_1997 RLGFLHTFGPTLIAGLIAGFATTSPHVRFVLEQGAAGSLDDLVATGDLDV
MAV_1538|M.avium_104 RLAYLSSFGSTVVPRLIARFKESSPRVTFTLEEGAAESIADRVRSGGVDI
MSMEG_0104|M.smegmatis_MC2_155 RLAFLHSQAGSFVPDLLRRFRAHAPHVQFSLTQGAAHDLLERLAGGQVDL
MMAR_4157|M.marinum_M ALGFLHSLGMQTVPHLIRLHHSRFPQARFELHQRSGQGVLSDLVSGVTDV
MUL_4020|M.ulcerans_Agy99 ALGFLHSLGMQTVPHLIRLHHSRFPQARFELHQRSGQGVLSDLVSGVTDV
Mb0121|M.bovis_AF2122/97 RLGTVPTASTTASLVLSAFCSAHPLAKVQVCSRLAATELYRRLREFELDA
Rv0117|M.tuberculosis_H37Rv RLGTVPTASTTASLVLSAFCSAHPLAKVQVCSRLAATELYRRLREFELDA
Mflv_3565|M.gilvum_PYR-GCK AIGTVT-LHPMNVAHLIAEFHADHPDVEITLGTATSDVLIAELGDGRLDL
Mvan_1027|M.vanbaalenii_PYR-1 VVGFTAASAHAVLPRLLERARERLPDVKLDLREMVTAAQIEGLMTGELDL
MLBr_02041|M.leprae_Br4923 RLGLIPTVAPYVLPTMLTGLTHRLPTLTLRVVEDQTEHLLTALREGALDA
TH_3504|M.thermoresistible__bu VIGHTPALSGAEAYALIEPAVTAYPSTSFTLRQLYPDALTDAVFDGTVQL
:. : : :
MAB_2707|M.abscessus_ATCC_1997 AIVSPRPRR--ANLGWRNLFRQRLGVAVPTDHRLAQAQ-DVSMVDLADEP
MAV_1538|M.avium_104 GVVSPRPVE--RTLAWRSLFRQRLGVAVPDGHRFARGA-AVSMVDLADEP
MSMEG_0104|M.smegmatis_MC2_155 AITSPRP----EGFQWRGLYVERLCLAVPRGHRLASRS-RITLADAGDEP
MMAR_4157|M.marinum_M SLSFPAAFEGYGEIAWQTLFTQQLYAVVSTDHPLAGRR-LIRFPELAAYP
MUL_4020|M.ulcerans_Agy99 SLSFPAAFEGYGEIAWQTLFTQQLYAVVSTDHPLAGRR-LIRFPELAAYP
Mb0121|M.bovis_AF2122/97 VIVHPETQDS-DDVDLVPLYEEQYVLLSPADMLPPGTS-TLVWRDAAQLP
Rv0117|M.tuberculosis_H37Rv VIVHPETQDS-DDVDLVPLYEEQYVLLSPADMLPPGTS-TLVWRDAAQLP
Mflv_3565|M.gilvum_PYR-GCK AIVSIAVDEDLPGLEVAVITDEAVEAAVADGHPLAHRR-SLSLEQLSRHP
Mvan_1027|M.vanbaalenii_PYR-1 GMARPPLKR--PGLVSRPLLHEQLLAALPEGHPLADMTRELTLIDLDGQD
MLBr_02041|M.leprae_Br4923 AMIALPVET--AGIAEIPIYDEDFVLALPPGHPLSGKR-RVPTTALAQLP
TH_3504|M.thermoresistible__bu GLRRGVVPT--PELATAVIGYHRVRVAFVDSHPLAARE-SVDITELAREQ
: . : . . . :
MAB_2707|M.abscessus_ATCC_1997 FVAMHPGFG--MRRLLEELCAAAQFQPRIVLE-SSNLMTVAGLVAAGLGV
MAV_1538|M.avium_104 FVTMHPGFG--MRRLLDELCAAAQFRPRVALQ-APNLTTAASLVAAGLGI
MSMEG_0104|M.smegmatis_MC2_155 FVGLAPGFG--LRQLTEELCAEAGITPHVVFE-ATEIPTMEGLVAAGFGV
MMAR_4157|M.marinum_M FVVLDRDHT--LRRIVDAACARHGIDPTLAFE-GTDVGTLRGLIGANLGV
MUL_4020|M.ulcerans_Agy99 FVVLDRDHT--LRRIVDAACARHGIDPTPAFE-GTDVGTLRGLIGANLGV
Mb0121|M.bovis_AF2122/97 LALLTADMR--DRQVIDAAFADHAVSAIPQVE-TDSVASLFAQVATGNWA
Rv0117|M.tuberculosis_H37Rv LALLTADMR--DRQVIDAAFADHAVSAIPQVE-TDSVASLFAQVATGNWA
Mflv_3565|M.gilvum_PYR-GCK LISLPAGTG--LRTRLDTACAAAGLRPRIAFE-ATSPAELAELARHGLGV
Mvan_1027|M.vanbaalenii_PYR-1 FVMYSPVQARYFNELLISTFTIAGTTPRYVQY-VTQVHTMLVLVRSGIGI
MLBr_02041|M.leprae_Br4923 LLLLDEGHC--LRDQTLDICRKSGVQAELANTRAASLATAVQCVTGGLGV
TH_3504|M.thermoresistible__bu VALWAPPGTSYYSDFLMGACRRAGFEPDYVVSRVQGAATVAAPLTTG-GI
. . .
MAB_2707|M.abscessus_ATCC_1997 TVLP------VDAT-TYPPDLRMLRLIDADAHR--DVGIVWSSGQPLSRP
MAV_1538|M.avium_104 SLVP------IDGS-SYPSGVSVKPLADADAYR--DVGMIWDSGRPLPRS
MSMEG_0104|M.smegmatis_MC2_155 AVVP------VPHAGRAEPGAEYIALAQASAKR--QLGLAWPADRPLPPA
MMAR_4157|M.marinum_M GVLP------QATA--RPADIIEIAIDDAQLVR--PIAIGWIADRCLPSS
MUL_4020|M.ulcerans_Agy99 GVLP------QATA--RPADIIEIAIDDAQLVR--PIAIGWIADRCLPSS
Mb0121|M.bovis_AF2122/97 SIVPHTWLWAMPMSGPTGGEIRAVELVDPVLKA--QIALATNALGPGSPV
Rv0117|M.tuberculosis_H37Rv SIVPHTWLWAMPMSGPTGGEIRAVELVDPVLKA--QIALATNALGPGSPV
Mflv_3565|M.gilvum_PYR-GCK AILP-------QSMARGGSGLHALRLAPELRGR--LVWAWRADGANPAAR
Mvan_1027|M.vanbaalenii_PYR-1 ALVP-------ASAATLHPEGVVFRTIGAFRERPVELDAVWRGDSTNPAL
MLBr_02041|M.leprae_Br4923 TLLP----QSAAPVESVRSKLGLAQFAAPCPGR--RIGLAFRSASGRSAS
TH_3504|M.thermoresistible__bu AFVT------AAAGPTMNGRVQVVDLTPPLLVP---VQALWQRHTLSAIR
.:. : .
MAB_2707|M.abscessus_ATCC_1997 VRNFISHAVAST----------------
MAV_1538|M.avium_104 ARDFIAGAAAVGHGGW------------
MSMEG_0104|M.smegmatis_MC2_155 AERFAQFVIRTTHDVE------------
MMAR_4157|M.marinum_M AVAFRETALGCYGHRVSAAGV-------
MUL_4020|M.ulcerans_Agy99 AVAFRETAFGCYGHRDSAAGV-------
Mb0121|M.bovis_AF2122/97 ARALITCAQALALNEFFDTQLRGITRRR
Rv0117|M.tuberculosis_H37Rv ARALITCAQALALNEFFDTQLRGITRRR
Mflv_3565|M.gilvum_PYR-GCK VLGERARAMAVSPGSGAPAVVPGA----
Mvan_1027|M.vanbaalenii_PYR-1 LRVLKDVLPQQEWTT-------------
MLBr_02041|M.leprae_Br4923 YRQIAKIIGELISTEHHVRLVA------
TH_3504|M.thermoresistible__bu DRILEGLG--------------------