For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSNTQLTDVIFILLLLVGFAQLLGYLFVKMRQPKVVGEILAGVVLGPALLGRLPFMEHIVDAVKHSGNIL TFVYWLGLLLLMFLSGAEMQQLFTRGERRTVGWLIIVGTGTPFLIGLIVGPHLVGPSLAGPHGNRLSLII ILAVGVAVTSVPVVSKIFADLNILDTRFARLVLGVAVLEDIVLWLALAIATAIATKTTLNAWQMSLHLAV TVAYFAVGLTILPVVMKRINKARWNAFAQQSPTGYAIAVLLAYCAVAGALDVNLVFAAFLAGFAVVHRKR RIFEDALEAIGKVSFAFFIPVYFAIVGLKLDLIRGVSLPMIAIFVLGTCTIKVMSVSLAGRFAGFRGLDL INLAVTTNARGGPGIVLASVAFDAGIISPKFYTTLVVAAVLTSQFAGAWLDYVLRKGWPLLASDNTKQAA QAAGADVSAGVMVASGQDSDVAEQPQEPVADPAPQRNTR
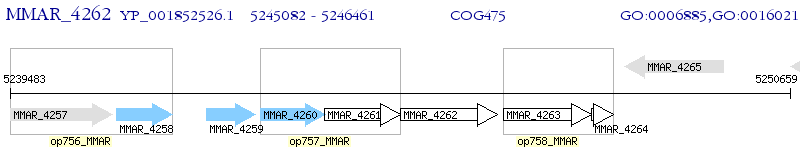
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4262 | - | - | 100% (459) | hypothetical protein MMAR_4262 |
| M. marinum M | MMAR_3635 | - | 1e-34 | 28.25% (400) | integral membrane ion antiporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3264c | - | 4e-05 | 21.22% (410) | integral membrane transport protein |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv3236c | - | 4e-05 | 21.22% (410) | integral membrane transport protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2285 | - | 3e-38 | 28.83% (444) | Na+/H+ antiporter |
| M. avium 104 | MAV_4118 | - | 2e-09 | 23.45% (388) | transporter, monovalent cation:proton antiporter-2 (CPA2) |
| M. smegmatis MC2 155 | MSMEG_6769 | - | 1e-27 | 23.88% (402) | transporter, monovalent cation:proton antiporter-2 (CPA2) |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1424 | - | 4e-11 | 22.94% (401) | sodium/hydrogen exchanger |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3264c|M.bovis_AF2122/97 -------------MEVSRALLFELGVLLAVLAVLG---AVARRFALSPIP
Rv3236c|M.tuberculosis_H37Rv -------------MEVSRALLFELGVLLAVLAVLG---AVARRFALSPIP
MAB_2285|M.abscessus_ATCC_1997 MSNTAVTAAAALSKDVSLIFFADLVILLVAALACG---AVAGKIGLPKMV
MSMEG_6769|M.smegmatis_MC2_155 -----------MPADTAIHFFLQLAVILMACRLAG---LVARRIGQAQVV
MMAR_4262|M.marinum_M -----------MSNTQLTDVIFILLLLVGFAQLLG---YLFVKMRQPKVV
MAV_4118|M.avium_104 -----------MQNERVQGFGFHTLALLTAVGFAGPLLASAKRFRIPVVI
Mvan_1424|M.vanbaalenii_PYR-1 ---------------MDGHLFPTIAIILALATAAG---VVATRLRQPLIV
. :: * :: . :
Mb3264c|M.bovis_AF2122/97 VYLLAGLSLG-----------NGGILGVAAAG-EFIATGAPIGVVLLLLA
Rv3236c|M.tuberculosis_H37Rv VYLLAGLSLG-----------NGGILGVAAAG-EFIATGAPIGVVLLLLA
MAB_2285|M.abscessus_ATCC_1997 GEIAAGVLLGPTVFGRLFPETKDALFPSAGTGSEIISGLCLLAVVLLVGV
MSMEG_6769|M.smegmatis_MC2_155 GEMIAGVILGPSLLGRIAPDLQGLLFPPGITN-VVLYTTAQIGLVLYMFI
MMAR_4262|M.marinum_M GEILAGVVLGPALLGRLP--FMEHIVDAVKHSGNILTFVYWLGLLLLMFL
MAV_4118|M.avium_104 GELIAGLAIG------------RTGFGVVDVADPTFQLLANIGFALVMFV
Mvan_1424|M.vanbaalenii_PYR-1 AFIMVGVAVG-------------PVGAGWVSADSTMELLARLGLAVLLFL
: .*: :* : :.. : :
Mb3264c|M.bovis_AF2122/97 LGLEFSATEFASSLRHHLPSAGVDIVLNATPG--AVAGWLLGLDG-----
Rv3236c|M.tuberculosis_H37Rv LGLEFSATEFASSLRHHLPSAGVDIVLNATPG--AVAGWLLGLDG-----
MAB_2285|M.abscessus_ATCC_1997 AGMHVDVDKLKQRLAGIATISVLGLAVPLAGG--MAVGYLVPEPLRPAAV
MSMEG_6769|M.smegmatis_MC2_155 IGLNFDVNHVKQRAGTAAAVSATGTLAPLALGGVAAIPLLAHGGFFGDGV
MMAR_4262|M.marinum_M SGAEMQQLFTRGERRTVGWLIIVGTGTPFLIG-LIVGPHLVGPSLAGPHG
MAV_4118|M.avium_104 VGTHVPLRARLMRSALPAALARAALAGGIAAVLGVALAVQFDTGH-----
Mvan_1424|M.vanbaalenii_PYR-1 VGLRLDLHMIRNTGPVAVITGLGQVLVTFALGLLIALALGMDN-------
* .. .
Mb3264c|M.bovis_AF2122/97 ---VAILGLAGVTYISSSGVIARLLEDLRRLGNRETPAVLSVLVLEDFAM
Rv3236c|M.tuberculosis_H37Rv ---VAILGLAGVTYISSSGVIARLLEDLRRLGNRETPAVLSVLVLEDFAM
MAB_2285|M.abscessus_ATCC_1997 DRTVFALFLGTAVAVSAIPVIARILMDLGLASHEIAQVILAAAIVDDVAG
MSMEG_6769|M.smegmatis_MC2_155 NVGMAMMFLGASVAITAFPMLARIIFEKRLSGTSLGTLALACGATSDAIS
MMAR_4262|M.marinum_M NRLSLIIILAVGVAVTSVPVVSKIFADLNILDTRFARLVLGVAVLEDIVL
MAV_4118|M.avium_104 -----AALYAVLMASSSAALALPVIDSLRLRGPR-VLSVTTQIAIADAAC
Mvan_1424|M.vanbaalenii_PYR-1 ---VTALYVAVALTFSSTIIIVKLLSDKRELEQLHGRIALGILIVQDIVV
. :: : :: . *
Mb3264c|M.bovis_AF2122/97 AAYLPLFAVLATD--GSWLEAVVGMTVAIAALLGAFAASYR--WGHHVGR
Rv3236c|M.tuberculosis_H37Rv AAYLPLFAVLATD--GSWLEAVVGMTVAIAALLGAFAASYR--WGHHVGR
MAB_2285|M.abscessus_ATCC_1997 WLLLSAVTGMASDS-GTGGVAVLKMAVAAIVAVVALTALTRPALRNRLAR
MSMEG_6769|M.smegmatis_MC2_155 WCILAVVLAVYRN---SPVMAVVAIVGGLVYTLVLLTLGRRAFAKLGDAA
MMAR_4262|M.marinum_M WLALAIATAIATKTTLNAWQMSLHLAVTVAYFAVGLTILPVVMKRINKAR
MAV_4118|M.avium_104 IVLLPLVIDIRRAP--TAALGALAVAGCAAALFVLLRAADRKGWRRRLHA
Mvan_1424|M.vanbaalenii_PYR-1 VVVMIALTASGRGGEGSVGAGIAAVALKGIGLLATLWVLMRFVLPWLMPQ
: . :. :
Mb3264c|M.bovis_AF2122/97 LVTHPDSEQLLLRVLGITLIVAAVAESLHASAAVGAFLVGLTLTGET-AD
Rv3236c|M.tuberculosis_H37Rv LVTHPDSEQLLLRVLGITLIVAAVAESLHASAAVGAFLVGLTLTGET-AD
MAB_2285|M.abscessus_ATCC_1997 RCSRWSAPTSTTVTVATIIVCATAGQLLGFEPVIGAFFAGILVIAAT-PA
MSMEG_6769|M.smegmatis_MC2_155 EARQAITAPMLSTVLIVLMACAWLTDTIGIYAIFGAFILGAAMPSG--FF
MMAR_4262|M.marinum_M WN-AFAQQSPTGYAIAVLLAYCAVAGALDVNLVFAAFLAGFAVVHRKRRI
MAV_4118|M.avium_104 YSEQHRFALELRTSLLVLFALAALAVATQVSIMLAGFAVGLVVGAVG--E
Mvan_1424|M.vanbaalenii_PYR-1 IARSQELLVLFG--VAYAVSVASFSEWLGFSSEVGAFLAGVSLAGTQ---
: . . ...* * :
Mb3264c|M.bovis_AF2122/97 RARMVLTP---LRDLFATIFFLGIGLSVDPGKLVS--MLPVALALAAVTA
Rv3236c|M.tuberculosis_H37Rv RARMVLTP---LRDLFATIFFLGIGLSVDPGKLVS--MLPVALALAAVTA
MAB_2285|M.abscessus_ATCC_1997 GSRVHKPLHAMAVAFFGPIFFATIGLQLDLGRLLSGPVLALTASICVVAV
MSMEG_6769|M.smegmatis_MC2_155 AERLTGRLEPLTTTFLLPLFFVYSGLNTEIGLVNTPFLWAVTAGLLVVAV
MMAR_4262|M.marinum_M FEDALEAIGKVSFAFFIPVYFAIVGLKLDLIRGVS---LPMIAIFVLGTC
MAV_4118|M.avium_104 PHRLARQLFGITEGFFSPLFFVWLGASLQVR--------ELGAHPQLILL
Mvan_1424|M.vanbaalenii_PYR-1 FRDALGARLVSLRDFLLFFFFLNLGAGLDFSEAAG--QITAAIILSVFVL
:: .:* * :
Mb3264c|M.bovis_AF2122/97 ATKVATGMFAARREGVARRGQLRAGTALVARGEFSLIIIGLAG----ASI
Rv3236c|M.tuberculosis_H37Rv ATKVATGMFAARREGVARRGQLRAGTALVARGEFSLIIIGLAG----ASI
MAB_2285|M.abscessus_ATCC_1997 FSKFLGAGMGARISGFSWPESVAVGAGLNTRGVIQIVIAMVGVRCGVFSP
MSMEG_6769|M.smegmatis_MC2_155 VGKGVACAVAARLSRVPVRDSLALGSLMNARGLIELILLNIGLEAGVITP
MMAR_4262|M.marinum_M TIKVMSVSLAGRFAGFRGLDLINLAVTTNARGGPGIVLASVAFDAGIISP
MAV_4118|M.avium_104 GAGLGCGAVLAHCAGRLLGQPLTLAVLSAAQLGVPVAAATIGTQQHLLAP
Mvan_1424|M.vanbaalenii_PYR-1 VAKPAIVIAIMSVMRYPVRVGFLVGLPLAQISEFSLILAALGLSLGHISN
. . : :. :
Mb3264c|M.bovis_AF2122/97 PGVAALATAYVFVMAIVGPILARYTGGGLPAAAVASN-------------
Rv3236c|M.tuberculosis_H37Rv PGVAALATAYVFVMAIVGPILARYTGGGLPAAAVASN-------------
MAB_2285|M.abscessus_ATCC_1997 EVFTALVVTTIATTAIAGPVLKRTLTPRRQLPAVISKPPQEPALNR----
MSMEG_6769|M.smegmatis_MC2_155 TLFTILVLVAIVTTLMASPIFEFVYGRHRTDETGDTTQERVPQPSA----
MMAR_4262|M.marinum_M KFYTTLVVAAVLTSQFAGAWLDYVLRKGWPLLASDNTKQAAQAAGADVSA
MAV_4118|M.avium_104 GEASALMFGALLTIAAASIAAGLAARRQGAPEPGEPAR------------
Mvan_1424|M.vanbaalenii_PYR-1 ATVSLITVVGLITIAASTYMTMYSDGIFARSQRWLSHLERKHQRDHEVAG
: : : .
Mb3264c|M.bovis_AF2122/97 --------------------------------------------------
Rv3236c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2285|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6769|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4262|M.marinum_M GVMVASGQDSDVAEQPQEPVADPAPQRNTR--------------------
MAV_4118|M.avium_104 --------------------------------------------------
Mvan_1424|M.vanbaalenii_PYR-1 LSDGDFDIILFGLGRFGSHLADRLSGAGHRVLGVDFDPHGVRAHRRDGVT
Mb3264c|M.bovis_AF2122/97 --------------------------------------------------
Rv3236c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2285|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6769|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4262|M.marinum_M --------------------------------------------------
MAV_4118|M.avium_104 --------------------------------------------------
Mvan_1424|M.vanbaalenii_PYR-1 TTFGSAEDLDLLEALPLDRAKYVISAIPVLQTNLALLHGLRQHQFSGAVA
Mb3264c|M.bovis_AF2122/97 --------------------------------------------------
Rv3236c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2285|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6769|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4262|M.marinum_M --------------------------------------------------
MAV_4118|M.avium_104 --------------------------------------------------
Mvan_1424|M.vanbaalenii_PYR-1 LTAHTRHDAERLRATGVEIVIEPFSSAAHTTSNTLHDLLTTEFEDTDDEG
Mb3264c|M.bovis_AF2122/97 --------------
Rv3236c|M.tuberculosis_H37Rv --------------
MAB_2285|M.abscessus_ATCC_1997 --------------
MSMEG_6769|M.smegmatis_MC2_155 --------------
MMAR_4262|M.marinum_M --------------
MAV_4118|M.avium_104 --------------
Mvan_1424|M.vanbaalenii_PYR-1 YSDDEGGAQPRPSR