For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MARSRITDWHPEDTVAWANGNYAIARRNLTWLVLNAHIAFSVWYLWSVMVLFMPQSIYGFTTGDKLLLDA TASLVGALVRIPYAMAANWFGGSTWTTISSLVLLIPTLGTIVLLAHPGLPLWPYLVCAAFTGLGGGNYSG SLAKVDGLFPQRLKGFVLGLTGGMANLGSAAVQMVGLVVLATAGHAAPYWVCAIYLVLLTIGGIGAALFM DDVVAHRTGLSLNNLRSILTVPDSWTISLLYMCASGSFLGFAFAFGQVLQHNFAAAGETQAHASLHAAQI AFIGPLLGSLARIVGGKLSDRFGGGHVTLTVFSAAILAGGFLVGVSIYADAAEERGDPVTGFMMAGYIVG FIALFIVCGAGKGAVYKLIPSVFEERSRGLSLNAAQRHHWARVRSGALIGFAGSFGALGGVGINMALRQS YVSTGTETPAFWIFFACYIGAAILTWVRYVRPTETAAGAAGRPAPLPASAPQPSAESAAEPTGAESITT
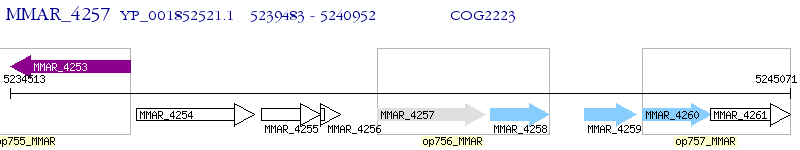
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4257 | narK3_3 | - | 100% (489) | nitrite extrusion protein, NarK3_3 |
| M. marinum M | MMAR_4265 | narK3_4 | 0.0 | 69.30% (469) | integral membrane nitrite extrusion protein NarK3_4 |
| M. marinum M | MMAR_2079 | narK3 | e-177 | 65.52% (464) | integral membrane nitrite extrusion protein NarK3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0267c | narK3 | 1e-158 | 58.00% (469) | integral membrane nitrite extrusion protein NarK3 |
| M. gilvum PYR-GCK | Mflv_3015 | - | 1e-120 | 48.31% (474) | nitrite extrusion protein |
| M. tuberculosis H37Rv | Rv0261c | narK3 | 1e-158 | 58.00% (469) | integral membrane nitrite extrusion protein NarK3 |
| M. leprae Br4923 | MLBr_00844 | narK | 1e-144 | 51.65% (484) | putative nitrite extrusion protein |
| M. abscessus ATCC 19977 | MAB_3523c | - | 1e-124 | 48.81% (461) | integral membrane nitrite extrusion protein NarK3 |
| M. avium 104 | MAV_2079 | - | 0.0 | 67.01% (479) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_0433 | - | 1e-162 | 57.89% (475) | nitrite extrusion protein |
| M. thermoresistible (build 8) | TH_0644 | narK2 | 1e-06 | 23.96% (192) | POSSIBLE NITRATE/NITRITE TRANSPORTER NARK2 |
| M. ulcerans Agy99 | MUL_0927 | narK3_3 | 0.0 | 96.26% (481) | nitrite extrusion protein, NarK3_3 |
| M. vanbaalenii PYR-1 | Mvan_3492 | - | 6e-61 | 33.63% (455) | nitrite transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3015|M.gilvum_PYR-GCK ------------------------------MTWAFTGVMAKRTRDIDTWD
MAB_3523c|M.abscessus_ATCC_199 -----------------------------MANNTATAPIARRGRWIENWD
MMAR_4257|M.marinum_M ---------------------------------------MARSR-ITDWH
MUL_0927|M.ulcerans_Agy99 ---------------------------------------MARSR-ITDWH
MAV_2079|M.avium_104 ---------------------------------------MARTRRIAHWD
Mb0267c|M.bovis_AF2122/97 ---------------------------------------MGRSHQISDWD
Rv0261c|M.tuberculosis_H37Rv ---------------------------------------MGRSHQISDWD
MSMEG_0433|M.smegmatis_MC2_155 ------------------------------------MPTLLKPKRITNWD
MLBr_00844|M.leprae_Br4923 MDQLVLLQAEESLYGPGRRSKTVPIDHRHSLKSSFFRLRLRRACRISHWD
Mvan_3492|M.vanbaalenii_PYR-1 ------------------MTVTDSESTGALATAAPAPERHRGKHWIDDWR
TH_0644|M.thermoresistible__bu --------------------------------------------------
Mflv_3015|M.gilvum_PYR-GCK PEDVQAWEGTGGKAPGKEIAKRNLIWSIVAEHVGFSVWSIWSVMVLFMPQ
MAB_3523c|M.abscessus_ATCC_199 PEDVVAWD-NG----GAKIARRNLIWSVVAEHVGFSVWSIWSVMVLFMPQ
MMAR_4257|M.marinum_M PEDTVAWANGN-----YAIARRNLTWLVLNAHIAFSVWYLWSVMVLFMPQ
MUL_0927|M.ulcerans_Agy99 PEDTVAWANGN-----YAIARRNLTWLVLNAHIAFSVWYLWSVMVLFMPQ
MAV_2079|M.avium_104 PEDLVAWEAGN-----KLIARRNLIWSIATMHVAFSIWYLWSVMVLFMPQ
Mb0267c|M.bovis_AF2122/97 PEDSVAWEAGN-----KFIARRNLIWSVAAEHVGFSVWSLWSVMVLFMPT
Rv0261c|M.tuberculosis_H37Rv PEDSVAWEAGN-----KFIARRNLIWSVAAEHVGFSVWSLWSVMVLFMPT
MSMEG_0433|M.smegmatis_MC2_155 PEDVAAWEAGN-----KYVARRNLIWSVVAEHIGFSIWSIWSVMVLFMPE
MLBr_00844|M.leprae_Br4923 PEDQAAWEAGN-----KTIARLNLLWSVLTVHLGYSVWTLWPVMELFMPK
Mvan_3492|M.vanbaalenii_PYR-1 PEDPEFWATTG-----KPVARRNLIFSIFAEHIGFSVWLLWSIVVVQMTA
TH_0644|M.thermoresistible__bu -----------------MIGPLSTTYAVDIS-------------------
:. . : :
Mflv_3015|M.gilvum_PYR-GCK DVYG--------IDAAGKFYLVAVPTLVGAFMRIPYTIAPARFGGRNWTI
MAB_3523c|M.abscessus_ATCC_199 NVYH--------IDAAGKFFLVAMPTLVGAVLRLPYTFATAWFGGRNWTV
MMAR_4257|M.marinum_M SIYG--------FTTGDKLLLDATASLVGALVRIPYAMAANWFGGSTWTT
MUL_0927|M.ulcerans_Agy99 SIYG--------FTTGDKLLLDATASLVGALVRIPYAMAANWFGGSTWTT
MAV_2079|M.avium_104 ARYG--------FTTGDKLLVGATAALVGALVRIPYAMGTARLGGRNWAV
Mb0267c|M.bovis_AF2122/97 SVYG--------FSAGDKFLLGATATLVGACLRFPYTFATAKFGGRNWTI
Rv0261c|M.tuberculosis_H37Rv SVYG--------FSAGDKFLLGATATLVGACLRFPYTFATAKFGGRNWTI
MSMEG_0433|M.smegmatis_MC2_155 AVYG--------FSAGDKFLLGATATLVGSCLRIPYTLATATFGGRNWTV
MLBr_00844|M.leprae_Br4923 DVYG--------FSAGDKFLLATTATLVGACLRVPYSLATALFGGRNWAI
Mvan_3492|M.vanbaalenii_PYR-1 GADGSPAASGFALTTTQALWLVAVPSGVGAFLRLPYTFAVPVFGGRNWTV
TH_0644|M.thermoresistible__bu ------------LSSAEASMLVATPILVGAVGRIVVGSLTDRYGGRAMFI
: : : : . **: *. **
Mflv_3015|M.gilvum_PYR-GCK VSALLLLIPTVLAYYYMKNPAS-YTTYMVIAAFAGLGGGNFASSMTNINA
MAB_3523c|M.abscessus_ATCC_199 FSALVLAVPTALTLYFMAHPETSYTTFMIVAAVAGFGGGNFASSMTNINA
MMAR_4257|M.marinum_M ISSLVLLIPTLGTIVLLAHPGLPLWPYLVCAAFTGLGGGNYSGSLAKVDG
MUL_0927|M.ulcerans_Agy99 ISSLVLLIPTLGTIVLLAHPGLPLWLYLVCAAFTGLGGGNYSGSLAKVDG
MAV_2079|M.avium_104 LSSLVLLIPTLAAIVLLAHPGLPLWPYLVCAALTGLGGGNYAAALANVES
Mb0267c|M.bovis_AF2122/97 FSALVLLIPTVGSILLLANPGLPLWPYLVCGALAGLGGGNFAASMTNINA
Rv0261c|M.tuberculosis_H37Rv FSALVLLIPTVGSILLLANPGLPLWPYLVCGALAGLGGGNFAASMTNINA
MSMEG_0433|M.smegmatis_MC2_155 FSALVLLIPTVMTVVLMANPGLPLWPYLVCAALAGFGGGNFASSMTNINA
MLBr_00844|M.leprae_Br4923 FSVMVLLIPTIATMVLLAHPGLPLWPYLACAALTGLGGGNFAASMTNANA
Mvan_3492|M.vanbaalenii_PYR-1 ISALLLLIPCLGLAWAVSNPDINFAVLLLIAATAGFGGGNFASSMANISF
TH_0644|M.thermoresistible__bu AITLASIVPVLAVG--AAGAAGSYPLLLVFGFFLGIAGTIFAVGIPFANN
: :* . : . *:.* :: .:. .
Mflv_3015|M.gilvum_PYR-GCK FYPQRLKGWALGLNAGGGNIGVPVIQLIGLMVIAAVGNTRPEIVCAIYLV
MAB_3523c|M.abscessus_ATCC_199 FYPQRFKGWALGLNAGGGNIGVPVIQVIGLLVIATVGNRSPHWVAAIYLV
MMAR_4257|M.marinum_M LFPQRLKGFVLGLTGGMANLGSAAVQMVGLVVLATAGHAAPYWVCAIYLV
MUL_0927|M.ulcerans_Agy99 LFPQRLKGFVLGLTGGMANLGSAAVQMVGLVVLATAGHAAPYRVCAIYLV
MAV_2079|M.avium_104 FFPQRRKGFALGLTGGVGNLGAAGIQAVGLVVLATAGNQAPYWVCAVYLV
Mb0267c|M.bovis_AF2122/97 FFPQRLKGAALALNAGGGNLGVPMVQLVGLLVIATAGDREPYWVCAIYLV
Rv0261c|M.tuberculosis_H37Rv FFPQRLKGAALALNAGGGNLGVPMVQLVGLLVIATAGDREPYWVCAIYLV
MSMEG_0433|M.smegmatis_MC2_155 FYPQRLKGWALGLNAGGGNIGVPMIQLVGLFVIATAGNRAPYWVCAVYLV
MLBr_00844|M.leprae_Br4923 FYPHRLKGAAFGLAGGAGNLGVSVIQVVGMLVIASVGDRKPYIVCGLYVV
Mvan_3492|M.vanbaalenii_PYR-1 FYPESEKGWALGLNAAGGNIGVAVAQKIIPVVVTLGAGVALSLAGLFYIP
TH_0644|M.thermoresistible__bu WYAPSRRGFATGVFG-MGMVGTALSAFFTPRFVESFGLFTTHVIIAVALG
:. :* . .: . . :* . . .: . . :
Mflv_3015|M.gilvum_PYR-GCK AISGAAVGAALFMDNLRNQK--SNFGALV----EAMRFKHSWVMSFLYIG
MAB_3523c|M.abscessus_ATCC_199 LIAFAAVGAALFMDNLADQK--TDGRSLI----DVMKYRDSWIIAFLYIG
MMAR_4257|M.marinum_M LLTIGGIGAALFMDDVVAHRTGLSLNNLR----SILTVPDSWTISLLYMC
MUL_0927|M.ulcerans_Agy99 LLTIGGIGAALFMDDVVAHRTGLSLNNLR----SILTVPDSWTISLLYMC
MAV_2079|M.avium_104 LLALVGAGAALFMDNLDHS---VEVGHVR----SVLTVPDTWTISFLYMC
Mb0267c|M.bovis_AF2122/97 LLAVAGLGAALYMDNLTEYR--IELNTMR----AVVSEPHTWVISLLYIG
Rv0261c|M.tuberculosis_H37Rv LLAVAGLGAALYMDNLTEYR--IELNTMR----AVVSEPHTWVISLLYIG
MSMEG_0433|M.smegmatis_MC2_155 ALAVAGLGAALYMDNLEHHK--IDLGAMK----AILGERDTWIISLLYIG
MLBr_00844|M.leprae_Br4923 LLIIAGIGAMLFMNDIEHHR--IGVNTIRPVLFVVVSTRDSWVLALLYLA
Mvan_3492|M.vanbaalenii_PYR-1 FTVAAAVCAFLFMNNLTEAK--ADVKPVW----ESLRHADTWIMSLLYIG
TH_0644|M.thermoresistible__bu LTALLCLVAMHDSPDFTPNT-ERVLPKLK----AAARLPVTWEMSILYAI
* :. : :* :::**
Mflv_3015|M.gilvum_PYR-GCK TFGSFIGFSFAFGQVLQINYLAG--------GDTPAQASLHAAQIAFLGP
MAB_3523c|M.abscessus_ATCC_199 TFGSFIGFSFAFGQVLQLNFLAGLAHSGVTPAQAAAQASLHAAQIAFIGP
MMAR_4257|M.marinum_M ASGSFLGFAFAFGQVLQHNFAAA--------GETQAHASLHAAQIAFIGP
MUL_0927|M.ulcerans_Agy99 ASGSFLGFAFAFGQVLQHNFAAA--------GETQAHASLHAAQIAFIGP
MAV_2079|M.avium_104 ASGSFIGFAFAFGQVIAHNLIAG--------GQTHGQAALHAAEIAFAGP
Mb0267c|M.bovis_AF2122/97 TFGSFIGFSFAFGQVLQINFIAS--------GQSTAQASLHAAQIAFLGP
Rv0261c|M.tuberculosis_H37Rv TFGSFIGFSFAFGQVLQINFIAS--------GQSTAQASLHAAQIAFLGP
MSMEG_0433|M.smegmatis_MC2_155 TFGSFIGFAFAFGQVLQINFISG--------GQTPAQASLHAAQIAFIGP
MLBr_00844|M.leprae_Br4923 SFGSFIGFSFAFGQVLETNFVAG--------GQSTAQAALHAAELAFIGP
Mvan_3492|M.vanbaalenii_PYR-1 TFGSFIGYSAAFPTLLKAVFDRG----------------DIALMWAFLGA
TH_0644|M.thermoresistible__bu VFGGFVAFSNYLPTYXPSSPACT---------------------------
*.*:.:: :
Mflv_3015|M.gilvum_PYR-GCK LLGSISRPFGGKLADRVGGGRITLYVFVAMIF-AAGVLVATGVMDDAA--
MAB_3523c|M.abscessus_ATCC_199 LLGSLARPFGGWLSDRTGGGKISLYAFIAMIF-GAGILVAAGTISDRG--
MMAR_4257|M.marinum_M LLGSLARIVGGKLSDRFGGGHVTLTVFSAAIL-AGGFLVGVSIYADAAEE
MUL_0927|M.ulcerans_Agy99 LLGSLARNVGGKLSDRFGGGHVTLTVFSAAIL-AGGFLVGVSIYADEAEE
MAV_2079|M.avium_104 LLGSMARVVGGKLGDRFDGGRVTLTVLAAMIV-AGGFLVAVSTHDDLTRP
Mb0267c|M.bovis_AF2122/97 LLGSLSRIYGGKLADRIGGGRVTLAAFCAMLL-ATGILISASTFGDHL--
Rv0261c|M.tuberculosis_H37Rv LLGSLSRIYGGKLADRIGGGRVTLAAFCAMLL-ATGILISASTFGDHL--
MSMEG_0433|M.smegmatis_MC2_155 LLGSLSRVYGGKLADRIGGGRVTLAVFAGMIL-GAGLLVAVSTVSDGS--
MLBr_00844|M.leprae_Br4923 TLAAVARFWGGRLADRLGGSRVTLVVFGAMVF-AAGLLGVLGIIEGSR--
Mvan_3492|M.vanbaalenii_PYR-1 GIGSVARPLGGKLSDRVGGARITAVSFAMLAIGAAGALWS----------
TH_0644|M.thermoresistible__bu -ITTCTSTPPPRHSTRFGSG------------------------------
: : : . * ...
Mflv_3015|M.gilvum_PYR-GCK -AGAPTSAQMIGYVAGFILLFILSGLGNGSTYKMIPSIFEAKAQGRDDLD
MAB_3523c|M.abscessus_ATCC_199 -AGAPSGSIMTAYVIGFIILFVVSGIGNGSVYKMIPSVFAAKARSAG-LD
MMAR_4257|M.marinum_M RGDPVTGFMMAGYIVGFIALFIVCGAGKGAVYKLIPSVFEERSRGLS-LN
MUL_0927|M.ulcerans_Agy99 RGDPVSGFMMAGYIVGFIALFIVCGAGKGAVYKLIPSVFEERSRGLS-LN
MAV_2079|M.avium_104 SGAPVSLYTTAGYIAGFIALFICCGVGKGSVFKLIPSVFAQRSRALE-LG
Mb0267c|M.bovis_AF2122/97 -AGPMPTATMVGYVIGFTALFILSGIGNGSVYKMIPSIFEARSHSLQ-IS
Rv0261c|M.tuberculosis_H37Rv -AGPMPTATMVGYVIGFTALFILSGIGNGSVYKMIPSIFEARSHSLQ-IS
MSMEG_0433|M.smegmatis_MC2_155 -SEAATGLMMSGYVVGFIALFLLSGIGNGSVYKMIPSIFEARSHSLD-VS
MLBr_00844|M.leprae_Br4923 -VCPIRGVMMASYFAGFITLFILSGLGNGSVYKMIPTIFEACSHSLG-IN
Mvan_3492|M.vanbaalenii_PYR-1 ----VQAKNMPVFFIAFMFLFVATGIGNGSTYRMISKIFRVKGEDAG--G
TH_0644|M.thermoresistible__bu ----------------------PGRSGPAGSWPMCSPPRNPVTTAGS---
* .. : : .
Mflv_3015|M.gilvum_PYR-GCK KEGKAAWSRAMSGALIGFAGAVGALGGVFINVVLRASYVGDAKSATAAFW
MAB_3523c|M.abscessus_ATCC_199 ET----WSRTMSGALIGVAGAIGALGGVAINLVLRASYLSAAKSATMAFW
MMAR_4257|M.marinum_M AAQRHHWARVRSGALIGFAGSFGALGGVGINMALRQSYVSTG-TETPAFW
MUL_0927|M.ulcerans_Agy99 AAQRHHWARVRSGALIGFAGSFGALGGVGINMALRQSYVSTG-TETPAFW
MAV_2079|M.avium_104 DTERRHWERARSAALIGFAGSFGALGGVVINLALRQSYASTG-SSTPAFG
Mb0267c|M.bovis_AF2122/97 EAERRQWSRSMSGALIGLAGAVGALGGVGVNLALRESYLTSG-TATSAFW
Rv0261c|M.tuberculosis_H37Rv EAERRQWSRSMSGALIGLAGAVGALGGVGVNLALRESYLTSG-TATSAFW
MSMEG_0433|M.smegmatis_MC2_155 EDERRHWSRSMSGALIGFAGAVGALGGVGINMALRQSYLSSG-SATTAFW
MLBr_00844|M.leprae_Br4923 DDECRDWSRVISGVVIGFVAEVGALGGVGIDLALRESYLNTG-GVTAAFW
Mvan_3492|M.vanbaalenii_PYR-1 DPLTMLHMRRQAAGALGIISSVGAFGGFIVPLAYAWSKSQFG-NIQPALQ
TH_0644|M.thermoresistible__bu --GRSAITRRSRGHWIGVRSTR------------SHPMCRCGCSTAAACC
* . :*. . . . *
Mflv_3015|M.gilvum_PYR-GCK VFLAFYVVCAFVTWFVFLRLQEH------------------RAHSGEHIG
MAB_3523c|M.abscessus_ATCC_199 VFLVFYVICAAVTWYAYVRRPMT------------------VSVP--AIS
MMAR_4257|M.marinum_M IFFACYIGAAILTWVRYVRPTETAAGAAGRPAPLPASAP-QPSAESAAEP
MUL_0927|M.ulcerans_Agy99 IFFACYIGAAILTWVRYVRPTETAAGAAGRPLHFRHPHPSRRQSRPPNQP
MAV_2079|M.avium_104 AFMLCYVAAAALTWARYVRPRRAPE----------------PRTAPDWQP
Mb0267c|M.bovis_AF2122/97 AFGVFYLVASVLTWAIYVRRGLK------------------SAGELVPAT
Rv0261c|M.tuberculosis_H37Rv AFGVFYLVASVLTWAIYVRRGLK------------------SAGELVPAT
MSMEG_0433|M.smegmatis_MC2_155 TFLAFYVLASVVTWFCYVRR---------------------PAAMPVTGT
MLBr_00844|M.leprae_Br4923 IFMLCYAAAGVLTWKMYVCRPLPGN-------------LHDEAANAFAAS
Mvan_3492|M.vanbaalenii_PYR-1 FYVGFFIVLLGVTWFFYMRKGTR---------------------------
TH_0644|M.thermoresistible__bu GR------------------------------------------------
Mflv_3015|M.gilvum_PYR-GCK RTSAPVPVG---------------
MAB_3523c|M.abscessus_ATCC_199 SSKNEVAV----------------
MMAR_4257|M.marinum_M TGAESITT----------------
MUL_0927|M.ulcerans_Agy99 RRSRSRPELDRGVAYPMPPSRGGE
MAV_2079|M.avium_104 MAEQPVSS----------------
Mb0267c|M.bovis_AF2122/97 TAPAGLAYV---------------
Rv0261c|M.tuberculosis_H37Rv TAPAGLAYV---------------
MSMEG_0433|M.smegmatis_MC2_155 ERHAPAIAG---------------
MLBr_00844|M.leprae_Br4923 VGASRTHRG---------------
Mvan_3492|M.vanbaalenii_PYR-1 MAQAGV------------------
TH_0644|M.thermoresistible__bu ------------------------