For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STPGEGSASSFYLPRLEYSTLPLAADRAVGWNTLRDAGPVVFMNGHYYLTRREDVLAALRNPKIFSSRIL QPPGNPLPVVPLAFDPPEHTRYRKALQPYFSPHALGKSRPVLERHAAEMIAGFAGRGECDAMAEFARLYP FQVFLDLYGLPLQDRDRVIAWKDAIIGDKLYLTREELEVGQLELLEYLNNAIQQRRQNPGSDMLSQVMTG PGEFTDLELLGMSHLLILAGLDTVTAAIGFSLLELARRPQLRTQLRDNPKEIRVFIEEIVRLEPSAPVAP RITTDFVNVGGMTLPPGSQVRLCMAAVNRDGSDAMSTNELVMDGKVHRHWGFGGGPHRCLGSHLARIELT VIVAEWLKQLPDFELAPDYSPEIAFPSKTFALKSLPLRWA*
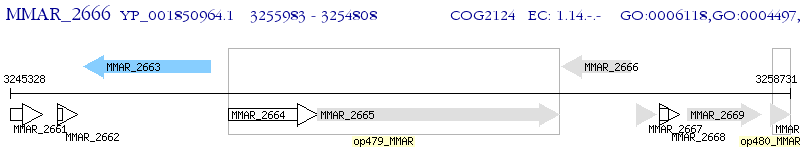
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2666 | cyp143A4 | - | 100% (391) | cytochrome P450 143A4 Cyp143A4 |
| M. marinum M | MMAR_2631 | cyp143A3 | e-126 | 58.05% (379) | cytochrome P450 143A3 Cyp143A3 |
| M. marinum M | MMAR_0274 | cyp271A1 | 7e-43 | 32.93% (331) | cytochrome P450 271A1 Cyp271A1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1813c | cyp143 | 1e-176 | 75.89% (394) | cytochrome P450 143 |
| M. gilvum PYR-GCK | Mflv_1764 | - | 1e-122 | 54.95% (384) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1785c | cyp143 | 1e-176 | 75.89% (394) | cytochrome P450 143 |
| M. leprae Br4923 | MLBr_02088 | - | 1e-30 | 27.99% (343) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1406c | - | 8e-34 | 30.98% (368) | cytochrome P450 |
| M. avium 104 | MAV_2931 | - | 0.0 | 78.44% (385) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_0762 | - | 2e-37 | 32.04% (362) | cytochrome P450 FAS1 |
| M. thermoresistible (build 8) | TH_3303 | - | 1e-40 | 32.05% (365) | PUTATIVE putative cytochrome P450 superfamily protein |
| M. ulcerans Agy99 | MUL_3091 | cyp143A4 | 0.0 | 99.74% (391) | cytochrome P450 143A4 Cyp143A4 |
| M. vanbaalenii PYR-1 | Mvan_4983 | - | 1e-121 | 56.20% (379) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2666|M.marinum_M ----------------------MSTPGEGSASSFYLPRLEYSTLPLAADR
MUL_3091|M.ulcerans_Agy99 ----------------------MSTPGEGSASSFYLPRLEYSTLPLSADR
MAV_2931|M.avium_104 ----------------------MS---EGQYGTFHLPRLDFATLPMSVDR
Mb1813c|M.bovis_AF2122/97 ----------------------MTTPGEDHAGSFYLPRLEYSTLPMAVDR
Rv1785c|M.tuberculosis_H37Rv ----------------------MTTPGEDHAGSFYLPRLEYSTLPMAVDR
Mflv_1764|M.gilvum_PYR-GCK ----------------------MTAT----AAPLGLPQVLYGDLPMSRDR
Mvan_4983|M.vanbaalenii_PYR-1 ----------------------MSA----------LPHVLYGDLPMAADR
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
MSMEG_0762|M.smegmatis_MC2_155 ------------------------------MTQAQALPPLHIRR-DAFDP
MAB_1406c|M.abscessus_ATCC_199 --------------------------MSVDVSHTVAPRYVPGTADNWTNP
TH_3303|M.thermoresistible__bu ---------------------------MTTFDTRPTVDFDHHSPEYAQNS
:
MMAR_2666|M.marinum_M AVGWNTLRDAGPVVFMNG-------HYYLTRREDVLAALRNPKIFSSR-I
MUL_3091|M.ulcerans_Agy99 AVGWNTLRDAGPVVFMNG-------HYYLTRREDVLAALRNPKIFSSR-I
MAV_2931|M.avium_104 GLGWKTLRDAGPVVFMNG-------HYYLTRREDVLAALRNPKVFSST-V
Mb1813c|M.bovis_AF2122/97 GVGWKTLRDAGPVVFMNG-------WYYLTRREDVLAALRNPKVFSSRKA
Rv1785c|M.tuberculosis_H37Rv GVGWKTLRDAGPVVFMNG-------WYYLTRREDVLAALRNPKVFSSRKA
Mflv_1764|M.gilvum_PYR-GCK GHGWATLRDLGPVLYGDG-------WFYLTRREDVLAALRNPEVFSSRIA
Mvan_4983|M.vanbaalenii_PYR-1 GAGWATLREFGPVAYGDG-------WYYLTRRDDVLAALRNPEVFSSRIA
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPGMPLT--VFSSFSDCDEALRHPLSASDRLKATLA
MSMEG_0762|M.smegmatis_MC2_155 TPELGEIRAGEGVHVTVNPFG--MQVYLVTRHEDVKTVLSDHERFSNSRP
MAB_1406c|M.abscessus_ATCC_199 WPMYAALRDHDPVHHVVPEYTPDQDYYVLTRHADVYEAARDWETFSSARG
TH_3303|M.thermoresistible__bu AAINAELRAKCPVAHTDAHGG----FWVISRYDDVVAAAKNDEVFASGYT
: : : . :
MMAR_2666|M.marinum_M LQPPGNPLP------------VVPLAFDPPEHTRYRKALQPYFSPHALGK
MUL_3091|M.ulcerans_Agy99 LQPPGNPLP------------VVPLAFDPPEHTRYRKALQPYFSPHALGK
MAV_2931|M.avium_104 LQPPGHPLP------------VLPLAFDPPQHTRYRKILQPYFSPHALGK
Mb1813c|M.bovis_AF2122/97 LQPPGNPLP------------VVPLAFDPPEHTRYRRILQPYFSPAALSK
Rv1785c|M.tuberculosis_H37Rv LQPPGNPLP------------VVPLAFDPPEHTRYRRILQPYFSPAALSK
Mflv_1764|M.gilvum_PYR-GCK YDDMISPVP------------LVPLGFDPPEHTRYRHILHPFFSPQTLAA
Mvan_4983|M.vanbaalenii_PYR-1 YDDMISPVP------------LVPLGFDPPEHTRYRRILHPFFSPQALGV
MLBr_02088|M.leprae_Br4923 QQAIAAGA------EPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQA
MSMEG_0762|M.smegmatis_MC2_155 PGFVLPGAPQISAEEQASNRAGNLLGLDPPEHQRLRRMLTPEFTIRRIKR
MAB_1406c|M.abscessus_ATCC_199 LTVTYGDLEKTG-----MGDNPPMVMQDPPTHTEFRKLVSRGFTPRQVTA
TH_3303|M.thermoresistible__bu VNGVSPLGVTIP----PSPVPMCPIEMDPPEYLPYRKLLNPFFSPGESKK
: *** : *: : *:
MMAR_2666|M.marinum_M SRPVLERHAAEMIAGFAGRG-ECDAMAEFARLYPFQVFLDLYGLPLQDRD
MUL_3091|M.ulcerans_Agy99 SRPVLERHAAEMIAGFAGRG-ECDAMAEFARLYPFQVFLDLYGLPLQDRD
MAV_2931|M.avium_104 SRPVLERHAAEMIAALADRG-ECEVMADFAHLYPFQVFMDLYGLPLQDRD
Mb1813c|M.bovis_AF2122/97 ALPSLRRHTVAMIDAIAGRG-ECEAMADLANLFPFQLFLVLYGLPLEDRD
Rv1785c|M.tuberculosis_H37Rv ALPSLRRHTVAMIDAIAGRG-ECEAMADLANLFPFQLFLVLYGLPLEDRD
Mflv_1764|M.gilvum_PYR-GCK LLPSLQAQVADIVEEIARRD-RCEVMAELATPYPSQVFLTLFGLPLQDRE
Mvan_4983|M.vanbaalenii_PYR-1 LLPSLQAQAADIVETIAQRD-ACEVMAELATPYPSQVFLTLFGLPLQDRE
MLBr_02088|M.leprae_Br4923 LEGDIAALVDSLLDKGAAAG-QFDVIADLAFPLAVAVICRLLGVPYEDAP
MSMEG_0762|M.smegmatis_MC2_155 LEPRIVEIVDAHLDAMESAGPPADIVADFALPIPSLVICELLGVPYEDRT
MAB_1406c|M.abscessus_ATCC_199 VEPKVREFVVQRLERLKERG-EGDIVVELFKPLPSMVVAHYLGVPEEDRT
TH_3303|M.thermoresistible__bu WEPRIAHWVDVCLDQVIEKG-SFDLVDDLANPVPSLFTCEFLGLPIEKWN
: . : . : : :: . . *:* :.
MMAR_2666|M.marinum_M RVIAWKDAIIG--DKLYL-------TREELEVGQLELLEYLNNAIQQRRQ
MUL_3091|M.ulcerans_Agy99 RVIAWKDAIIG--DKLYL-------TREELEVGQLELLEYLNNAIQQRRQ
MAV_2931|M.avium_104 RLLDWKNAVVG--EKPFV-------TESDVEK-SEQLLAYLADAIAQRRQ
Mb1813c|M.bovis_AF2122/97 RLIGWKDAVIAMSDRPHP-------TEADVAA-ARELLEYLTAMVAERRR
Rv1785c|M.tuberculosis_H37Rv RLIGWKDAVIAMSDRPHP-------TEADVAA-ARELLEYLTAMVAERRR
Mflv_1764|M.gilvum_PYR-GCK RLIAWKDAIIAFSLTTDP-------SSVDLKP-AVELYTYLSEAVAAQRD
Mvan_4983|M.vanbaalenii_PYR-1 RLIAWKDAIIAFSLTNDP-------TSVDLTP-AVELYGYLTEAVARQRE
MLBr_02088|M.leprae_Br4923 EFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRG
MSMEG_0762|M.smegmatis_MC2_155 DFQQRSARQLDLSAPMP-----------ERLELQRQGRAYMRGLVERSRT
MAB_1406c|M.abscessus_ATCC_199 QFDAWTDGIVAAASGGAID---IEAMQGEVAQTIGELMMYFTGLIERRRA
TH_3303|M.thermoresistible__bu DFASVQHEIIYTPPEEQE----------DILRRYMELCGEVFQLITERRQ
. : : . : *
MMAR_2666|M.marinum_M NP-GSDMLS---QVMTGPG--EFTDLELLGMSHLLILAGLDTVTAAIGFS
MUL_3091|M.ulcerans_Agy99 NP-GSDMLS---QVMTGPG--EFTDLELLGMSHLLILAGLDTVTAAIGFS
MAV_2931|M.avium_104 HP-GTDMLS---QVMTGEG--NFTDIELLGMSHLLILAGLDTVTAAIGFS
Mb1813c|M.bovis_AF2122/97 NP-GPDVLS---QVQIGED--PLSEIEVLGLSHLLILAGLDTVTAAVGFS
Rv1785c|M.tuberculosis_H37Rv NP-GPDVLS---QVQIGED--PLSEIEVLGLSHLLILAGLDTVTAAVGFS
Mflv_1764|M.gilvum_PYR-GCK DP-REGVLS---HLLHGDD--PLTDNEAVGLSLVFVLAGLDTVTSAIGAT
Mvan_4983|M.vanbaalenii_PYR-1 KP-REGILS---HLLHGDE--QLTDNEAVGLSLVFVLAGLDTVTSTIGAT
MLBr_02088|M.leprae_Br4923 TP-GEDLISRLIELDESGD--QLTEEEIIATCGLLLVAGHETTVNLIANA
MSMEG_0762|M.smegmatis_MC2_155 RP-GDDILG--MLVREHGT--ELTDDELIGIAGLLLLAGHETTSNMLGLG
MAB_1406c|M.abscessus_ATCC_199 EP-EDDTVSHLVAAGVGADGDISGTLQILGFAFTMVTGGNDTTTGMLGGA
TH_3303|M.thermoresistible__bu NPTGEGLIDFLVTAEIDGE--PIPDDKVFSMVNLVIAGGFDTTTAVTVSS
* . :. : .. .: .* :*.
MMAR_2666|M.marinum_M LLELARRPQLRTQLRDNPKEIRVFIEEIVRLEPSAPVAP-RITTDFVNVG
MUL_3091|M.ulcerans_Agy99 LLELARRPQLRTQLRDNPKEIRVFIEEIVRLEPSAPVAP-RITTDFVNVG
MAV_2931|M.avium_104 LFELARRPQLRKELRDNPKQTRVFIEEIVRLEPSAPVAP-RITTEYVEVG
Mb1813c|M.bovis_AF2122/97 LLELARRPQLRAMLRDNPKQIRVFIEEIVRLEPSAPVAP-RVTTEPVTVG
Rv1785c|M.tuberculosis_H37Rv LLELARRPQLRAMLRDNPKQIRVFIEEIVRLEPSAPVAP-RVTTEPVTVG
Mflv_1764|M.gilvum_PYR-GCK MLELARRPELRGALREDPEGIAVFVEEMIRLEPAAPIVG-RVTTRPVTVA
Mvan_4983|M.vanbaalenii_PYR-1 MLELARRPELRAALLGDPDGIAGFVEEMIRLEPAAPVVG-RVTTREVTVA
MLBr_02088|M.leprae_Br4923 VLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIG-RVAAKDMTIG
MSMEG_0762|M.smegmatis_MC2_155 VLALLRHPDQLACVRDDPDAVGPAIEELLRWLSIVSTALPRITTTDVELA
MAB_1406c|M.abscessus_ATCC_199 IQLLHQNPAQRQKLIDDPSLIPGAVEEFLRLTSPVQGLA-RTATRDVIIG
TH_3303|M.thermoresistible__bu LVYLAEHPEDRQRLIDNPDLLPRAIEEFLRAFTPQQALA-RTVTKPVTVA
: : ..* : :*. :** :* . * .: : :.
MMAR_2666|M.marinum_M GMTLPPGSQVRLCMAAVNRDGSDA-MSTNELVMDGKVHRHWGFGGGPHRC
MUL_3091|M.ulcerans_Agy99 GMTLPPGSQVRLCMAAVNRDGSDA-MSTNELVMDGKVHRHWGFGGGPHRC
MAV_2931|M.avium_104 GMTLPPGTSVRLCMAAVNRDDSDS-MSTNELNMDGKVHRHWGFGGGPHRC
Mb1813c|M.bovis_AF2122/97 GMTLPAGSPVRLCMAAVNRDGSDA-MSTDELVMDGKVHRHWGFGGGPHRC
Rv1785c|M.tuberculosis_H37Rv GMTLPAGSPVRLCMAAVNRDGSDA-MSTDELVMDGKVHRHWGFGGGPHRC
Mflv_1764|M.gilvum_PYR-GCK GVTLPAGSEVRLCLGAINRDGTDE-HSGDDLVLDGKLHKHWGFGGGPHRC
Mvan_4983|M.vanbaalenii_PYR-1 GVTLPKGAEVRLCLGAINRDGADE-HSGDDFVLDGKLHKHWGFGGGPHRC
MLBr_02088|M.leprae_Br4923 QTTLTEGDTMVLLLAAANRDPAVY-SRPDEFDPDRPSSRHLAFAVGSHFC
MSMEG_0762|M.smegmatis_MC2_155 GVTIPAGHLVFASLPAGNRDPEFI-DDPDTLDIRRGAPGHLAFGHGVHHC
MAB_1406c|M.abscessus_ATCC_199 DTTIPAGRRALLLYGSANRDEREYGNDAAELDVLRKPRNILTFSHGNHHC
TH_3303|M.thermoresistible__bu GVELQPGERVLLSWASANQDETAF-ERPEEIILDRFPNRHTTFGIGIHRC
: * : *:* : *. * * *
MMAR_2666|M.marinum_M LGSHLARIELTVIVAEWLKQLPDFELAPDYSPEIAFP-SKTFALKSLPLR
MUL_3091|M.ulcerans_Agy99 LGSHLARIELTVIVAEWLKQLPDFELAPDYSPEIAFP-SKTFALKSLPLR
MAV_2931|M.avium_104 LGSHLARIELTVVVAEWLTQIPDFELPDGYEPVINYP-SKSFALKELPLH
Mb1813c|M.bovis_AF2122/97 LGSHLARLELTLLVGEWLNQIPDFELAPDYAPEIRFP-SKSFALKNLPLR
Rv1785c|M.tuberculosis_H37Rv LGSHLARLELTLLVGEWLNQIPDFELAPDYAPEIRFP-SKSFALKNLPLR
Mflv_1764|M.gilvum_PYR-GCK LGSHLARMELKLVVSEWLSRIPDFDLEPGYVPEITWP-SATCTLTGLPLR
Mvan_4983|M.vanbaalenii_PYR-1 LGSHLARMELRLVVTEWLSRIPDFELAPGYVPEITWP-SATCTLPTLPLI
MLBr_02088|M.leprae_Br4923 LGAALARLEATVTLSAISARFPQVQLAG---ELVYKPNVAMRGMSALPVQ
MSMEG_0762|M.smegmatis_MC2_155 LGAPLARMEMRIALPALLRRFPTLALAEPFEDVRWRPFHFIYGLQSLAVA
MAB_1406c|M.abscessus_ATCC_199 LGAAAARMQSRIALEELLTRIPDFEVDLDGVTWADGS--YVRRPLTVPIR
TH_3303|M.thermoresistible__bu LGSHVARIELETMIRRVLDRMPDYHVDLAAARRYPSIGIINGWINAPATF
**: **:: : ::* : .
MMAR_2666|M.marinum_M WA------------
MUL_3091|M.ulcerans_Agy99 WA------------
MAV_2931|M.avium_104 WG------------
Mb1813c|M.bovis_AF2122/97 WS------------
Rv1785c|M.tuberculosis_H37Rv WS------------
Mflv_1764|M.gilvum_PYR-GCK ILR-----------
Mvan_4983|M.vanbaalenii_PYR-1 LTRKTP--------
MLBr_02088|M.leprae_Br4923 V-------------
MSMEG_0762|M.smegmatis_MC2_155 W-------------
MAB_1406c|M.abscessus_ATCC_199 VR------------
TH_3303|M.thermoresistible__bu TPGQRLRDEKLPGA