For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSALPHVLYGDLPMAADRGAGWATLREFGPVAYGDGWYYLTRRDDVLAALRNPEVFSSRIAYDDMISPVP LVPLGFDPPEHTRYRRILHPFFSPQALGVLLPSLQAQAADIVETIAQRDACEVMAELATPYPSQVFLTLF GLPLQDRERLIAWKDAIIAFSLTNDPTSVDLTPAVELYGYLTEAVARQREKPREGILSHLLHGDEQLTDN EAVGLSLVFVLAGLDTVTSTIGATMLELARRPELRAALLGDPDGIAGFVEEMIRLEPAAPVVGRVTTREV TVAGVTLPKGAEVRLCLGAINRDGADEHSGDDFVLDGKLHKHWGFGGGPHRCLGSHLARMELRLVVTEWL SRIPDFELAPGYVPEITWPSATCTLPTLPLILTRKTP
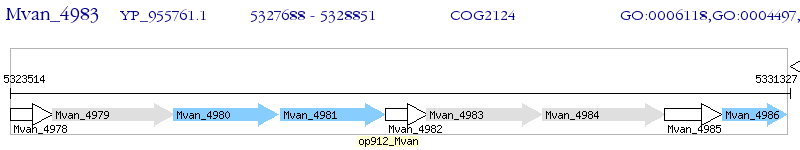
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_4983 | - | - | 100% (387) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_4465 | - | 6e-42 | 30.34% (379) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_5579 | - | 2e-41 | 32.82% (390) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1813c | cyp143 | 1e-131 | 59.42% (377) | cytochrome P450 143 |
| M. gilvum PYR-GCK | Mflv_1764 | - | 0.0 | 87.14% (381) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1785c | cyp143 | 1e-131 | 59.42% (377) | cytochrome P450 143 |
| M. leprae Br4923 | MLBr_02088 | - | 3e-34 | 29.89% (368) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1406c | - | 4e-40 | 30.93% (375) | cytochrome P450 |
| M. marinum M | MMAR_2631 | cyp143A3 | 1e-145 | 66.58% (380) | cytochrome P450 143A3 Cyp143A3 |
| M. avium 104 | MAV_2548 | - | 1e-131 | 59.79% (378) | putative cytochrome P450 143 |
| M. smegmatis MC2 155 | MSMEG_3551 | - | 3e-40 | 30.96% (365) | linalool 8-monooxygenase |
| M. thermoresistible (build 8) | TH_3303 | - | 7e-44 | 30.99% (355) | PUTATIVE putative cytochrome P450 superfamily protein |
| M. ulcerans Agy99 | MUL_3106 | cyp143A3 | 1e-144 | 66.32% (380) | cytochrome P450 143A3 Cyp143A3 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_4983|M.vanbaalenii_PYR-1 MSA----------LPHVLYGDLPMA----------------------ADR
Mflv_1764|M.gilvum_PYR-GCK MTATAAPLG----LPQVLYGDLPMS----------------------RDR
MMAR_2631|M.marinum_M MSAPRDERD----IPHFVYAELPMA----------------------TDR
MUL_3106|M.ulcerans_Agy99 MSAPRDERD----IPHFVYAELPMA----------------------TDR
MAV_2548|M.avium_104 MET----LG----APELSFASLPMA----------------------ADR
Mb1813c|M.bovis_AF2122/97 MTTPGEDHAGSFYLPRLEYSTLPMA----------------------VDR
Rv1785c|M.tuberculosis_H37Rv MTTPGEDHAGSFYLPRLEYSTLPMA----------------------VDR
MAB_1406c|M.abscessus_ATCC_199 MSVDVSHTV----APRYVPGTADNW----------------------TNP
MSMEG_3551|M.smegmatis_MC2_155 MGIAPRVNG--AAPPDVPLSEINLGSWD-------------FWALDDDIR
TH_3303|M.thermoresistible__bu MTTFDTRPT-----VDFDHHSPEYA----------------------QNS
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
*
Mvan_4983|M.vanbaalenii_PYR-1 GAGWATLREFGPVAYGD------------GWYYLTRRDDVLAALRN----
Mflv_1764|M.gilvum_PYR-GCK GHGWATLRDLGPVLYGD------------GWFYLTRREDVLAALRN----
MMAR_2631|M.marinum_M GRGWATLRDAGPVVIGD------------GFYYLTRRDDVLDALRH----
MUL_3106|M.ulcerans_Agy99 GRGWATLRDAGPVVIGD------------GFYYLTRRDDVLDALRH----
MAV_2548|M.avium_104 GVGWKVLRDAGRVVSVD------------GIFYLTHREDVLAALRD----
Mb1813c|M.bovis_AF2122/97 GVGWKTLRDAGPVVFMN------------GWYYLTRREDVLAALRN----
Rv1785c|M.tuberculosis_H37Rv GVGWKTLRDAGPVVFMN------------GWYYLTRREDVLAALRN----
MAB_1406c|M.abscessus_ATCC_199 WPMYAALRDHDPVHHVVPEYTPD-----QDYYVLTRHADVYEAARD----
MSMEG_3551|M.smegmatis_MC2_155 DGAFATLRREAPISFHPAFDTEEGFPQGAGHWALTRHDDVFYASRH----
TH_3303|M.thermoresistible__bu AAINAELRAKCPVAHTDAHG---------GFWVISRYDDVVAAAKN----
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPG-----------MPLTVFSSFSDCDEALRHPLSA
* : :: * * :.
Mvan_4983|M.vanbaalenii_PYR-1 PEVFSS---RIAYDDMISPVPLVPLG-------FDPPEHTRYRRILHPFF
Mflv_1764|M.gilvum_PYR-GCK PEVFSS---RIAYDDMISPVPLVPLG-------FDPPEHTRYRHILHPFF
MMAR_2631|M.marinum_M PEVFSS---KRAFDILGSPLPLVPLA-------FDPPEHTRFRKVLQPFF
MUL_3106|M.ulcerans_Agy99 PEVFSS---KRAFDILGSPLPLVPLA-------FDPPEHTRFRKVLQPFF
MAV_2548|M.avium_104 PELFSS---KKAFDVLGSPLPLVPIS-------FDPPEHTRFRKILQPFF
Mb1813c|M.bovis_AF2122/97 PKVFSS---RKALQPPGNPLPVVPLA-------FDPPEHTRYRRILQPYF
Rv1785c|M.tuberculosis_H37Rv PKVFSS---RKALQPPGNPLPVVPLA-------FDPPEHTRYRRILQPYF
MAB_1406c|M.abscessus_ATCC_199 WETFSS---ARGLTVTYGDLEKTGMGDNPPMVMQDPPTHTEFRKLVSRGF
MSMEG_3551|M.smegmatis_MC2_155 PELFSS---ASGIVIGDQTPELAEYFG--SMIAMDDPRHTRLRNIVRSAF
TH_3303|M.thermoresistible__bu DEVFASGYTVNGVSPLGVTIPPSPVP--MCPIEMDPPEYLPYRKLLNPFF
MLBr_02088|M.leprae_Br4923 SDRLKATLAQQAIAAGAEPRPFYASS----FMFLDPPDHTRLRKLVSKAF
. : : . * * : *.:: *
Mvan_4983|M.vanbaalenii_PYR-1 SPQALGVLLPSLQAQAADIVETIAQR---DACEVMAELATPYPSQVFLTL
Mflv_1764|M.gilvum_PYR-GCK SPQTLAALLPSLQAQVADIVEEIARR---DRCEVMAELATPYPSQVFLTL
MMAR_2631|M.marinum_M SPHALAAMLPSLQAQAIEIIDAIAAG---SQCEVITELAIPYPSQVFLTL
MUL_3106|M.ulcerans_Agy99 SPHALAAMLPSLQAQAIEIIDAIAAG---SQCEVITKLAIPYPSQVFLTL
MAV_2548|M.avium_104 SPHTLKEMLPSLQQQAIEIIERVAAQ---GECEVVADVAIPYPSQVFLTL
Mb1813c|M.bovis_AF2122/97 SPAALSKALPSLRRHTVAMIDAIAGR---GECEAMADLANLFPFQLFLVL
Rv1785c|M.tuberculosis_H37Rv SPAALSKALPSLRRHTVAMIDAIAGR---GECEAMADLANLFPFQLFLVL
MAB_1406c|M.abscessus_ATCC_199 TPRQVTAVEPKVREFVVQRLERLKER---GEGDIVVELFKPLPSMVVAHY
MSMEG_3551|M.smegmatis_MC2_155 TPRVLAVIEDSVRDRARRLVAGMVDRNPDGHAELVTELAGPLPLQIICDM
TH_3303|M.thermoresistible__bu SPGESKKWEPRIAHWVDVCLDQVIEK---GSFDLVDDLANPVPSLFTCEF
MLBr_02088|M.leprae_Br4923 APKVVQALEGDIAALVDSLLDKGAAA---GQFDVIADLAFPLAVAVICRL
:* : . : . : : .: . .
Mvan_4983|M.vanbaalenii_PYR-1 FGLPLQDRERLIAWKDAIIAFSL--------TNDPTSVDLTPAVELYGYL
Mflv_1764|M.gilvum_PYR-GCK FGLPLQDRERLIAWKDAIIAFSL--------TTDPSSVDLKPAVELYTYL
MMAR_2631|M.marinum_M FGLSLADRDRLIRWKDAIIALGL--------LASLDDADLTPALELTSYL
MUL_3106|M.ulcerans_Agy99 FGLSLADRDRLIRWKDAIIALGL--------LASLDDADLTPALELTSYL
MAV_2548|M.avium_104 FGLPLEDRDKLVAWKTSVIAISE--------APSLEDADLTPALELVAYL
Mb1813c|M.bovis_AF2122/97 YGLPLEDRDRLIGWKDAVIAMSD--------RPHPTEADVAAARELLEYL
Rv1785c|M.tuberculosis_H37Rv YGLPLEDRDRLIGWKDAVIAMSD--------RPHPTEADVAAARELLEYL
MAB_1406c|M.abscessus_ATCC_199 LGVPEEDRTQFDAWTDGIVAAASGGA---IDIEAMQGEVAQTIGELMMYF
MSMEG_3551|M.smegmatis_MC2_155 MGIPESDHQQIFHWTNVILGFGDP------DLTTDFDEFVTVAMDIGAYA
TH_3303|M.thermoresistible__bu LGLPIEKWNDFASVQHEIIYTPP----------EEQEDILRRYMELCGEV
MLBr_02088|M.leprae_Br4923 LGVPYEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYL
*:. . : :: :
Mvan_4983|M.vanbaalenii_PYR-1 TEAVARQREKPREGILSHLLHG-----DEQLTDNEAVGLSLVFVLAGLDT
Mflv_1764|M.gilvum_PYR-GCK SEAVAAQRDDPREGVLSHLLHG-----DDPLTDNEAVGLSLVFVLAGLDT
MMAR_2631|M.marinum_M VDAIAAHRRHPGTDILSQVLLG-----DDPLDDSEALGLSYVFVLAGLDT
MUL_3106|M.ulcerans_Agy99 VDAIAAHRRHPGTDILSQVLLG-----DDPLDDSEALGLSYVFVLAGLDT
MAV_2548|M.avium_104 TEAINARRADPGPDILSQLLNG-----EEPLDDAEIMGLSFLFVLAGLDT
Mb1813c|M.bovis_AF2122/97 TAMVAERRRNPGPDVLSQVQIG-----EDPLSEIEVLGLSHLLILAGLDT
Rv1785c|M.tuberculosis_H37Rv TAMVAERRRNPGPDVLSQVQIG-----EDPLSEIEVLGLSHLLILAGLDT
MAB_1406c|M.abscessus_ATCC_199 TGLIERRRAEPEDDTVSHLVAAGVGADGDISGTLQILGFAFTMVTGGNDT
MSMEG_3551|M.smegmatis_MC2_155 TALADQRRSAPADDLTTSLVQAEVD--GERLTSAEVASFFILLVVAGNET
TH_3303|M.thermoresistible__bu FQLITERRQNPTGEGLIDFLVTAEID-GEPIPDDKVFSMVNLVIAGGFDT
MLBr_02088|M.leprae_Br4923 EQLVKCRRGTPGEDLISRLIELDES--GDQLTEEEIIATCGLLLVAGHET
:* * . : : . .: .* :*
Mvan_4983|M.vanbaalenii_PYR-1 VTSTIGATMLELARRPELRAALLGDPDGIA-GFVEEMIRLEPAAPVVGRV
Mflv_1764|M.gilvum_PYR-GCK VTSAIGATMLELARRPELRGALREDPEGIA-VFVEEMIRLEPAAPIVGRV
MMAR_2631|M.marinum_M VTSAIGAALLELARRPQLRQLLRENHGQIA-VFVDEIVRLEPPAPVVPRV
MUL_3106|M.ulcerans_Agy99 VTSAIGAALLELARRPQLRQLLRENHGQIA-VFVDEIVRLEPPAPVVPRV
MAV_2548|M.avium_104 VTAAMSSALLELARNPELRATLHDDPDQID-VFVEEIVRLEPPAPMLPRV
Mb1813c|M.bovis_AF2122/97 VTAAVGFSLLELARRPQLRAMLRDNPKQIR-VFIEEIVRLEPSAPVAPRV
Rv1785c|M.tuberculosis_H37Rv VTAAVGFSLLELARRPQLRAMLRDNPKQIR-VFIEEIVRLEPSAPVAPRV
MAB_1406c|M.abscessus_ATCC_199 TTGMLGGAIQLLHQNPAQRQKLIDDPSLIP-GAVEEFLRLTSPVQGLART
MSMEG_3551|M.smegmatis_MC2_155 TRNAISHGVLALTRYPEQRRLWWSRFDELAPTAVEEIVRWASPVSYMRRT
TH_3303|M.thermoresistible__bu TTAVTVSSLVYLAEHPEDRQRLIDNPDLLP-RAIEEFLRAFTPQQALART
MLBr_02088|M.leprae_Br4923 TVNLIANAVLAMLRNPSQWKALSSNPQRAP-LVVEETLRYDPAIHLIGRV
. : : . * ::* :* .. *.
Mvan_4983|M.vanbaalenii_PYR-1 TTREVTVAGVTLPKGAEVRLCLGAINRDGADE-HSGDDFVLDGKLHKHWG
Mflv_1764|M.gilvum_PYR-GCK TTRPVTVAGVTLPAGSEVRLCLGAINRDGTDE-HSGDDLVLDGKLHKHWG
MMAR_2631|M.marinum_M TTREVNIAGAKIPAGSRVWLCLGAINRDGSDA-TSVNDVVMDGKVHKHWG
MUL_3106|M.ulcerans_Agy99 TTREVNIAGAKIPAGSRVWLCLGAINRDGSDA-TSVNDVVMDGKVHKHWD
MAV_2548|M.avium_104 TTTEVTIGDVTLPADTMVRLCVAAINRDDSDE-ISTNDVVMDGKVHRHWG
Mb1813c|M.bovis_AF2122/97 TTEPVTVGGMTLPAGSPVRLCMAAVNRDGSDA-MSTDELVMDGKVHRHWG
Rv1785c|M.tuberculosis_H37Rv TTEPVTVGGMTLPAGSPVRLCMAAVNRDGSDA-MSTDELVMDGKVHRHWG
MAB_1406c|M.abscessus_ATCC_199 ATRDVIIGDTTIPAGRRALLLYGSANRDEREYGNDAAELDVLRKPRNILT
MSMEG_3551|M.smegmatis_MC2_155 VTRDTELAGTALPAGSKVTLWYGSANRDETKF-DNPWMFDVQRHPNPHVG
TH_3303|M.thermoresistible__bu VTKPVTVAGVELQPGERVLLSWASANQDETAF-ERPEEIILDRFPNRHTT
MLBr_02088|M.leprae_Br4923 AAKDMTIGQTTLTEGDTMVLLLAAANRDPAVY-SRPDEFDPDRPSSRHLA
.: :. : . * .: *:* .
Mvan_4983|M.vanbaalenii_PYR-1 FGGG-PHRCLGSHLARMELRLVVTEWLSRIPDFELAPGYVPEITWPSATC
Mflv_1764|M.gilvum_PYR-GCK FGGG-PHRCLGSHLARMELKLVVSEWLSRIPDFDLEPGYVPEITWPSATC
MMAR_2631|M.marinum_M FGGG-THRCLGSHLARMELSLVISEWLRRIPQFELACGYTPEIDWPSATL
MUL_3106|M.ulcerans_Agy99 FGGG-THRCLGSHLARMELSLVISEWLCRIPQFELACGYTPEIDWPSATL
MAV_2548|M.avium_104 FGGG-PHRCLGSHLARLELKLIVGEWLRRIPEFSVKVGEEPQIVFPASTF
Mb1813c|M.bovis_AF2122/97 FGGG-PHRCLGSHLARLELTLLVGEWLNQIPDFELAPDYAPEIRFPSKSF
Rv1785c|M.tuberculosis_H37Rv FGGG-PHRCLGSHLARLELTLLVGEWLNQIPDFELAPDYAPEIRFPSKSF
MAB_1406c|M.abscessus_ATCC_199 FSHG-NHHCLGAAAARMQSRIALEELLTRIPDFEVDLDG---VTWADGSY
MSMEG_3551|M.smegmatis_MC2_155 FGGGGAHFCLGANLARREITVLFSELHRHLP--DIVATEEPDRLQSAFIH
TH_3303|M.thermoresistible__bu FGIG-IHRCLGSHVARIELETMIRRVLDRMPDYHVDLAAARRYPSIGIIN
MLBr_02088|M.leprae_Br4923 FAVG-SHFCLGAALARLEATVTLSAISARFP--QVQLAGELVYKPNVAMR
*. * * ***: ** : . ::* :
Mvan_4983|M.vanbaalenii_PYR-1 TLPTLPLILTRKTP---------
Mflv_1764|M.gilvum_PYR-GCK TLTGLPLRILR------------
MMAR_2631|M.marinum_M GLSTLTLQFTS------------
MUL_3106|M.ulcerans_Agy99 GLSTLPLQFTS------------
MAV_2548|M.avium_104 SLERVPLKLG-------------
Mb1813c|M.bovis_AF2122/97 ALKNLPLRWS-------------
Rv1785c|M.tuberculosis_H37Rv ALKNLPLRWS-------------
MAB_1406c|M.abscessus_ATCC_199 VRRPLTVPIRVR-----------
MSMEG_3551|M.smegmatis_MC2_155 GIKRLPVAWR-------------
TH_3303|M.thermoresistible__bu GWINAPATFTPGQRLRDEKLPGA
MLBr_02088|M.leprae_Br4923 GMSALPVQV--------------
.