For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TATAAPLGLPQVLYGDLPMSRDRGHGWATLRDLGPVLYGDGWFYLTRREDVLAALRNPEVFSSRIAYDDM ISPVPLVPLGFDPPEHTRYRHILHPFFSPQTLAALLPSLQAQVADIVEEIARRDRCEVMAELATPYPSQV FLTLFGLPLQDRERLIAWKDAIIAFSLTTDPSSVDLKPAVELYTYLSEAVAAQRDDPREGVLSHLLHGDD PLTDNEAVGLSLVFVLAGLDTVTSAIGATMLELARRPELRGALREDPEGIAVFVEEMIRLEPAAPIVGRV TTRPVTVAGVTLPAGSEVRLCLGAINRDGTDEHSGDDLVLDGKLHKHWGFGGGPHRCLGSHLARMELKLV VSEWLSRIPDFDLEPGYVPEITWPSATCTLTGLPLRILR*
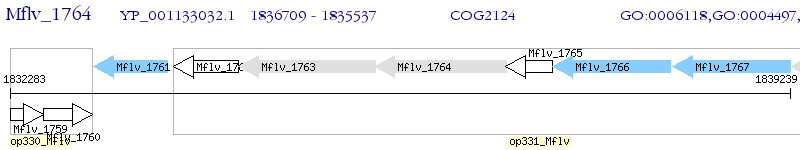
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1764 | - | - | 100% (390) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_2229 | - | 2e-38 | 30.00% (370) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_3307 | - | 1e-37 | 30.91% (372) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1813c | cyp143 | 1e-135 | 60.05% (378) | cytochrome P450 143 |
| M. tuberculosis H37Rv | Rv1785c | cyp143 | 1e-135 | 60.05% (378) | cytochrome P450 143 |
| M. leprae Br4923 | MLBr_02088 | - | 1e-34 | 29.46% (387) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1406c | - | 6e-39 | 29.95% (394) | cytochrome P450 |
| M. marinum M | MMAR_2631 | cyp143A3 | 1e-146 | 66.67% (378) | cytochrome P450 143A3 Cyp143A3 |
| M. avium 104 | MAV_2548 | - | 1e-136 | 59.84% (381) | putative cytochrome P450 143 |
| M. smegmatis MC2 155 | MSMEG_5038 | - | 4e-37 | 29.41% (357) | putative cytochrome P450 123 |
| M. thermoresistible (build 8) | TH_3303 | - | 1e-43 | 30.79% (354) | PUTATIVE putative cytochrome P450 superfamily protein |
| M. ulcerans Agy99 | MUL_3106 | cyp143A3 | 1e-146 | 66.40% (378) | cytochrome P450 143A3 Cyp143A3 |
| M. vanbaalenii PYR-1 | Mvan_4983 | - | 0.0 | 87.14% (381) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1813c|M.bovis_AF2122/97 MTTPGEDHAGSFYLPRLEYSTLPMAV----------------------DR
Rv1785c|M.tuberculosis_H37Rv MTTPGEDHAGSFYLPRLEYSTLPMAV----------------------DR
Mflv_1764|M.gilvum_PYR-GCK MTATAAPLG----LPQVLYGDLPMSR----------------------DR
Mvan_4983|M.vanbaalenii_PYR-1 MSA----------LPHVLYGDLPMAA----------------------DR
MMAR_2631|M.marinum_M MSAPRDERD----IPHFVYAELPMAT----------------------DR
MUL_3106|M.ulcerans_Agy99 MSAPRDERD----IPHFVYAELPMAT----------------------DR
MAV_2548|M.avium_104 MET----LG----APELSFASLPMAA----------------------DR
MAB_1406c|M.abscessus_ATCC_199 MSVDVSHT----VAPRYVPGTADNWT----------------------NP
MSMEG_5038|M.smegmatis_MC2_155 MGADLSHP----AVA-FELANADTWA----------------------DP
TH_3303|M.thermoresistible__bu MTTFDTRPT-----VDFDHHSPEYAQ----------------------NS
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
* . :
Mb1813c|M.bovis_AF2122/97 GVGWKTLRDAGPVVFMNG-------WYYLTRREDVLAALRNPKVFSS---
Rv1785c|M.tuberculosis_H37Rv GVGWKTLRDAGPVVFMNG-------WYYLTRREDVLAALRNPKVFSS---
Mflv_1764|M.gilvum_PYR-GCK GHGWATLRDLGPVLYGDG-------WFYLTRREDVLAALRNPEVFSS---
Mvan_4983|M.vanbaalenii_PYR-1 GAGWATLREFGPVAYGDG-------WYYLTRRDDVLAALRNPEVFSS---
MMAR_2631|M.marinum_M GRGWATLRDAGPVVIGDG-------FYYLTRRDDVLDALRHPEVFSS---
MUL_3106|M.ulcerans_Agy99 GRGWATLRDAGPVVIGDG-------FYYLTRRDDVLDALRHPEVFSS---
MAV_2548|M.avium_104 GVGWKVLRDAGRVVSVDG-------IFYLTHREDVLAALRDPELFSS---
MAB_1406c|M.abscessus_ATCC_199 WPMYAALRDHDPVHHVVPEYTPDQDYYVLTRHADVYEAARDWETFSS---
MSMEG_5038|M.smegmatis_MC2_155 WDMYTALRDHDPVHHVVPPERPHHDYFVLSRHADIWAAARDHETYSS---
TH_3303|M.thermoresistible__bu AAINAELRAKCPVAHTDAHG----GFWVISRYDDVVAAAKNDEVFASGYT
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPGMP------LTVFSSFSDCDEALRHPLSASDR-L
* : :: * * :. :.
Mb1813c|M.bovis_AF2122/97 -----RKALQPPGNPLPVVPLA-------FDPPEHTRYRRILQPYFSPAA
Rv1785c|M.tuberculosis_H37Rv -----RKALQPPGNPLPVVPLA-------FDPPEHTRYRRILQPYFSPAA
Mflv_1764|M.gilvum_PYR-GCK -----RIAYDDMISPVPLVPLG-------FDPPEHTRYRHILHPFFSPQT
Mvan_4983|M.vanbaalenii_PYR-1 -----RIAYDDMISPVPLVPLG-------FDPPEHTRYRRILHPFFSPQA
MMAR_2631|M.marinum_M -----KRAFDILGSPLPLVPLA-------FDPPEHTRFRKVLQPFFSPHA
MUL_3106|M.ulcerans_Agy99 -----KRAFDILGSPLPLVPLA-------FDPPEHTRFRKVLQPFFSPHA
MAV_2548|M.avium_104 -----KKAFDVLGSPLPLVPIS-------FDPPEHTRFRKILQPFFSPHT
MAB_1406c|M.abscessus_ATCC_199 -----ARGLTVTYGDLEKTGMGDNPPMVMQDPPTHTEFRKLVSRGFTPRQ
MSMEG_5038|M.smegmatis_MC2_155 -----AQGLTVNYGELELIGLADNPPFVMQDPPTHTEFRKLVSRGFTPRQ
TH_3303|M.thermoresistible__bu VNGVSPLGVTIPPSPVPMCPIE-------MDPPEYLPYRKLLNPFFSPGE
MLBr_02088|M.leprae_Br4923 KATLAQQAIAAGAEPRPFYASS----FMFLDPPDHTRLRKLVSKAFAPKV
. *** : *::: *:*
Mb1813c|M.bovis_AF2122/97 LSKALPSLRRHTVAMIDAIAGRGECEAMADLANLFPFQLFLVLYGLPLED
Rv1785c|M.tuberculosis_H37Rv LSKALPSLRRHTVAMIDAIAGRGECEAMADLANLFPFQLFLVLYGLPLED
Mflv_1764|M.gilvum_PYR-GCK LAALLPSLQAQVADIVEEIARRDRCEVMAELATPYPSQVFLTLFGLPLQD
Mvan_4983|M.vanbaalenii_PYR-1 LGVLLPSLQAQAADIVETIAQRDACEVMAELATPYPSQVFLTLFGLPLQD
MMAR_2631|M.marinum_M LAAMLPSLQAQAIEIIDAIAAGSQCEVITELAIPYPSQVFLTLFGLSLAD
MUL_3106|M.ulcerans_Agy99 LAAMLPSLQAQAIEIIDAIAAGSQCEVITKLAIPYPSQVFLTLFGLSLAD
MAV_2548|M.avium_104 LKEMLPSLQQQAIEIIERVAAQGECEVVADVAIPYPSQVFLTLFGLPLED
MAB_1406c|M.abscessus_ATCC_199 VTAVEPKVREFVVQRLERLKERGEGDIVVELFKPLPSMVVAHYLGVPEED
MSMEG_5038|M.smegmatis_MC2_155 VEAVEPQVREFVVERLERLRAEGGGDIVAELFKPLPSMVVAHYLGVPEAD
TH_3303|M.thermoresistible__bu SKKWEPRIAHWVDVCLDQVIEKGSFDLVDDLANPVPSLFTCEFLGLPIEK
MLBr_02088|M.leprae_Br4923 VQALEGDIAALVDSLLDKGAAAGQFDVIADLAFPLAVAVICRLLGVPYED
: . :: . : : .: . . *:. .
Mb1813c|M.bovis_AF2122/97 RDRL--------IGWKDAVIAMSDRPHPTEADVAAARELLE-----YLTA
Rv1785c|M.tuberculosis_H37Rv RDRL--------IGWKDAVIAMSDRPHPTEADVAAARELLE-----YLTA
Mflv_1764|M.gilvum_PYR-GCK RERL--------IAWKDAIIAFSLTTDPSSVDLKPAVELYT-----YLSE
Mvan_4983|M.vanbaalenii_PYR-1 RERL--------IAWKDAIIAFSLTNDPTSVDLTPAVELYG-----YLTE
MMAR_2631|M.marinum_M RDRL--------IRWKDAIIALGLLASLDDADLTPALELTS-----YLVD
MUL_3106|M.ulcerans_Agy99 RDRL--------IRWKDAIIALGLLASLDDADLTPALELTS-----YLVD
MAV_2548|M.avium_104 RDKL--------VAWKTSVIAISEAPSLEDADLTPALELVA-----YLTE
MAB_1406c|M.abscessus_ATCC_199 RTQF--------DAWTDGIVAAASGGAIDIEAMQGEVAQTIGELMMYFTG
MSMEG_5038|M.smegmatis_MC2_155 RAQF--------DGWTEAIVAANSEAGSALGAGDAVMSMTG-----YFAE
TH_3303|M.thermoresistible__bu WNDF--------ASVQHEIIYTP--PEEQEDILRRYMELCG-----EVFQ
MLBr_02088|M.leprae_Br4923 APEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRD-----YLEQ
: : .
Mb1813c|M.bovis_AF2122/97 MVAERRRNPGPDVLSQVQIG-----EDPLSEIEVLGLSHLLILAGLDTVT
Rv1785c|M.tuberculosis_H37Rv MVAERRRNPGPDVLSQVQIG-----EDPLSEIEVLGLSHLLILAGLDTVT
Mflv_1764|M.gilvum_PYR-GCK AVAAQRDDPREGVLSHLLHG-----DDPLTDNEAVGLSLVFVLAGLDTVT
Mvan_4983|M.vanbaalenii_PYR-1 AVARQREKPREGILSHLLHG-----DEQLTDNEAVGLSLVFVLAGLDTVT
MMAR_2631|M.marinum_M AIAAHRRHPGTDILSQVLLG-----DDPLDDSEALGLSYVFVLAGLDTVT
MUL_3106|M.ulcerans_Agy99 AIAAHRRHPGTDILSQVLLG-----DDPLDDSEALGLSYVFVLAGLDTVT
MAV_2548|M.avium_104 AINARRADPGPDILSQLLNG-----EEPLDDAEIMGLSFLFVLAGLDTVT
MAB_1406c|M.abscessus_ATCC_199 LIERRRAEPEDDTVSHLVAAGVGADGDISGTLQILGFAFTMVTGGNDTTT
MSMEG_5038|M.smegmatis_MC2_155 LIERRRREPGDDTISHLVAA--GADDEAVDVLSILAFTFTMVTGGNDTTT
TH_3303|M.thermoresistible__bu LITERRQNPTGEGLIDFLVTAEID-GEPIPDDKVFSMVNLVIAGGFDTTT
MLBr_02088|M.leprae_Br4923 LVKCRRGTPGEDLISRLIELD--ESGDQLTEEEIIATCGLLLVAGHETTV
: :* * : . : . .. .: .* :*..
Mb1813c|M.bovis_AF2122/97 AAVGFSLLELARRPQLRAMLRDNPKQIRVFIEEIVRLEPSAPVAPRVTTE
Rv1785c|M.tuberculosis_H37Rv AAVGFSLLELARRPQLRAMLRDNPKQIRVFIEEIVRLEPSAPVAPRVTTE
Mflv_1764|M.gilvum_PYR-GCK SAIGATMLELARRPELRGALREDPEGIAVFVEEMIRLEPAAPIVGRVTTR
Mvan_4983|M.vanbaalenii_PYR-1 STIGATMLELARRPELRAALLGDPDGIAGFVEEMIRLEPAAPVVGRVTTR
MMAR_2631|M.marinum_M SAIGAALLELARRPQLRQLLRENHGQIAVFVDEIVRLEPPAPVVPRVTTR
MUL_3106|M.ulcerans_Agy99 SAIGAALLELARRPQLRQLLRENHGQIAVFVDEIVRLEPPAPVVPRVTTR
MAV_2548|M.avium_104 AAMSSALLELARNPELRATLHDDPDQIDVFVEEIVRLEPPAPMLPRVTTT
MAB_1406c|M.abscessus_ATCC_199 GMLGGAIQLLHQNPAQRQKLIDDPSLIPGAVEEFLRLTSPVQGLARTATR
MSMEG_5038|M.smegmatis_MC2_155 GMLGGAVQLLQQHPDQRRVLTDDPDLIPDAVDEFLRLTSPVQGLARTATR
TH_3303|M.thermoresistible__bu AVTVSSLVYLAEHPEDRQRLIDNPDLLPRAIEEFLRAFTPQQALARTVTK
MLBr_02088|M.leprae_Br4923 NLIANAVLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAK
:: : ..* * : ::* :* .. *..:
Mb1813c|M.bovis_AF2122/97 PVTVGGMTLPAGSPVRLCMAAVNRDGSDA-MSTDELVMDGKVHRHWGFGG
Rv1785c|M.tuberculosis_H37Rv PVTVGGMTLPAGSPVRLCMAAVNRDGSDA-MSTDELVMDGKVHRHWGFGG
Mflv_1764|M.gilvum_PYR-GCK PVTVAGVTLPAGSEVRLCLGAINRDGTDE-HSGDDLVLDGKLHKHWGFGG
Mvan_4983|M.vanbaalenii_PYR-1 EVTVAGVTLPKGAEVRLCLGAINRDGADE-HSGDDFVLDGKLHKHWGFGG
MMAR_2631|M.marinum_M EVNIAGAKIPAGSRVWLCLGAINRDGSDA-TSVNDVVMDGKVHKHWGFGG
MUL_3106|M.ulcerans_Agy99 EVNIAGAKIPAGSRVWLCLGAINRDGSDA-TSVNDVVMDGKVHKHWDFGG
MAV_2548|M.avium_104 EVTIGDVTLPADTMVRLCVAAINRDDSDE-ISTNDVVMDGKVHRHWGFGG
MAB_1406c|M.abscessus_ATCC_199 DVIIGDTTIPAGRRALLLYGSANRDEREYGNDAAELDVLRKPRNILTFSH
MSMEG_5038|M.smegmatis_MC2_155 DVTIGDTTIPAGRKVLLLYGSGNRDEREFGPDAAQLDVRRRPRNILTFSH
TH_3303|M.thermoresistible__bu PVTVAGVELQPGERVLLSWASANQDETAF-ERPEEIILDRFPNRHTTFGI
MLBr_02088|M.leprae_Br4923 DMTIGQTTLTEGDTMVLLLAAANRDPAVY-SRPDEFDPDRPSSRHLAFAV
: :. : . * .: *:* :. . *.
Mb1813c|M.bovis_AF2122/97 GPHRCLGSHLARLELTLLVGEWLNQIPDFELAPDYAPEIRFPSKSFALKN
Rv1785c|M.tuberculosis_H37Rv GPHRCLGSHLARLELTLLVGEWLNQIPDFELAPDYAPEIRFPSKSFALKN
Mflv_1764|M.gilvum_PYR-GCK GPHRCLGSHLARMELKLVVSEWLSRIPDFDLEPGYVPEITWPSATCTLTG
Mvan_4983|M.vanbaalenii_PYR-1 GPHRCLGSHLARMELRLVVTEWLSRIPDFELAPGYVPEITWPSATCTLPT
MMAR_2631|M.marinum_M GTHRCLGSHLARMELSLVISEWLRRIPQFELACGYTPEIDWPSATLGLST
MUL_3106|M.ulcerans_Agy99 GTHRCLGSHLARMELSLVISEWLCRIPQFELACGYTPEIDWPSATLGLST
MAV_2548|M.avium_104 GPHRCLGSHLARLELKLIVGEWLRRIPEFSVKVGEEPQIVFPASTFSLER
MAB_1406c|M.abscessus_ATCC_199 GNHHCLGAAAARMQSRIALEELLTRIPDFEVDLD---GVTWADGSYVRRP
MSMEG_5038|M.smegmatis_MC2_155 GAHFCLGAAAARMQSRVALTELLTRCPDFTVDLD---AVVWAGGSYVRRP
TH_3303|M.thermoresistible__bu GIHRCLGSHVARIELETMIRRVLDRMPDYHVDLAAARRYPSIGIINGWIN
MLBr_02088|M.leprae_Br4923 GSHFCLGAALARLEATVTLSAISARFPQVQLAGELVYKPNVAMRGMSALP
* * ***: **:: : : *: :
Mb1813c|M.bovis_AF2122/97 LPLRWS-------------
Rv1785c|M.tuberculosis_H37Rv LPLRWS-------------
Mflv_1764|M.gilvum_PYR-GCK LPLRILR------------
Mvan_4983|M.vanbaalenii_PYR-1 LPLILTRKTP---------
MMAR_2631|M.marinum_M LTLQFTS------------
MUL_3106|M.ulcerans_Agy99 LPLQFTS------------
MAV_2548|M.avium_104 VPLKLG-------------
MAB_1406c|M.abscessus_ATCC_199 LTVPIRVR-----------
MSMEG_5038|M.smegmatis_MC2_155 LSVPFRAGA----------
TH_3303|M.thermoresistible__bu APATFTPGQRLRDEKLPGA
MLBr_02088|M.leprae_Br4923 VQV----------------