For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPQPTASSFAVLGLLGVQPWTAYELVAQSKRSLHWFWPRSERHLYAELKRLVERGHAEAHVEGGRRRRTR YTITPAGRAALQAWLGTEPAPPTLEIEGLLRVMLADQGNMDDLRAAVEATARQARTVRAEGLPLVAELLA TGGPFPQRLHLNERLVSFYSDFNNLLLRWCEETLADVETWQGTQDIGLTPSGRQRLVRAISEHP*
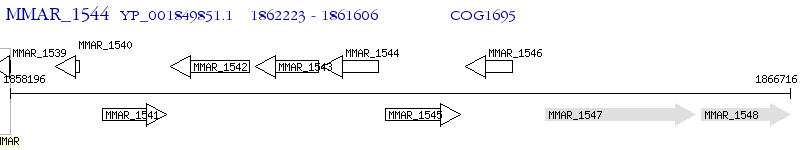
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1544 | - | - | 100% (205) | hypothetical protein MMAR_1544 |
| M. marinum M | MMAR_3454 | - | 2e-12 | 34.55% (110) | putative regulatory protein |
| M. marinum M | MMAR_0152 | - | 4e-12 | 27.47% (182) | hypothetical protein MMAR_0152 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1209c | - | 3e-06 | 32.06% (131) | hypothetical protein Mb1209c |
| M. gilvum PYR-GCK | Mflv_1331 | - | 3e-09 | 35.79% (95) | PadR-like family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1176c | - | 3e-06 | 32.06% (131) | hypothetical protein Rv1176c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0082c | - | 3e-11 | 34.25% (146) | hypothetical protein MAB_0082c |
| M. avium 104 | MAV_0135 | - | 2e-13 | 32.50% (200) | transcriptional regulator, PadR family protein |
| M. smegmatis MC2 155 | MSMEG_6226 | - | 2e-10 | 37.89% (95) | transcriptional regulator |
| M. thermoresistible (build 8) | TH_0324 | - | 5e-10 | 30.30% (132) | transcriptional regulator, PadR family protein |
| M. ulcerans Agy99 | MUL_2388 | - | 1e-116 | 98.54% (205) | hypothetical protein MUL_2388 |
| M. vanbaalenii PYR-1 | Mvan_5461 | - | 2e-13 | 37.39% (115) | PadR-like family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1544|M.marinum_M ---------MTPQPTASSFAVLGLLG-VQPWTAYELVAQSKRSLHWFWPR
MUL_2388|M.ulcerans_Agy99 ---------MTPQPTASSFAVLGLLG-VQPWTAYELVAQSKRSLHWFWPR
MAB_0082c|M.abscessus_ATCC_199 --MAAISSPGEPNLAATSWTILGMLSYEEEVSGYDIKKWADWSISYFYWS
TH_0324|M.thermoresistible__bu VTEAVEDAADQPNLSPTGWALLGMLSGGNELSGYDIKKWTDWVIRFFYSS
MSMEG_6226|M.smegmatis_MC2_155 --------------MSLRMAALGLLV-QQPGSGYDLLRRFEKSMANVWPA
Mvan_5461|M.vanbaalenii_PYR-1 --------------MSLRIAALGLLA-EGSASGYDLLKQFEKSMANVWPA
Mflv_1331|M.gilvum_PYR-GCK --MSIVECHRYYSSMSLRMAALGLLA-QHPASGYDLLKKFEVSMANVWPG
Mb1209c|M.bovis_AF2122/97 --------------MALPHAILVSLC-EQASSGYELARRFDRSIGYFWTA
Rv1176c|M.tuberculosis_H37Rv --------------MALPHAILVSLC-EQASSGYELARRFDRSIGYFWTA
MAV_0135|M.avium_104 --------------MSLRDAVLAALL-EGESSGYDLAKDFDASVANFWPA
. : * * :.*:: . : .:
MMAR_1544|M.marinum_M -SERHLYAELKRLVERGHAEAHVEGG--RRRRTRYTITPAGRAALQAWLG
MUL_2388|M.ulcerans_Agy99 -SERHLYAELKRLVERGHAEAHVEGG--RRRRTRYTITPAGRAALQAWLG
MAB_0082c|M.abscessus_ATCC_199 PSFSQVYSELRKLEKIGYATSRVDHDNGARSRRLYKITPAGMAAMSQWAN
TH_0324|M.thermoresistible__bu PAYSQIYSELKRLEQYGLVTSRIEGS--TRSRRMYKITDAGLAAVTRWTN
MSMEG_6226|M.smegmatis_MC2_155 -TQSQLYGELNKLAAAGLIEVSAVGP---RGRKEYRATDAGRAELLRWIT
Mvan_5461|M.vanbaalenii_PYR-1 -TQSQLYSELNKLAAGGLIEVCAVGP---RGRKEYRITDAGRAELQRWVA
Mflv_1331|M.gilvum_PYR-GCK -TQSQLYSELNKLADDGLIEVTAIGA---RGRKEYAITRPGRDELRRWVM
Mb1209c|M.bovis_AF2122/97 -THQQIYRTLRVMENNNWVRATTVLQHGRPDKKVYAISDSGRAELARWIA
Rv1176c|M.tuberculosis_H37Rv -THQQIYRTLRVMENNNWVRATTVLQHGRPDKKVYAISDSGRAELARWIA
MAV_0135|M.avium_104 -TPQQLYRELDRLAGQGLIRARVVHQQRRPNKRMFSLTAAGRAAIRRFTA
: ::* * : . : : : .* : :
MMAR_1544|M.marinum_M TEPAPP----------TLEIEGLLRVMLADQGNMDDLRAAVEATARQART
MUL_2388|M.ulcerans_Agy99 TEPAPP----------TLEIEGLVRVMLADQGNMDDLRAAVEATARQART
MAB_0082c|M.abscessus_ATCC_199 ETPAEPQ---------VLKHSVVLRVMQGHMTDPARLKEILERHIEYAEN
TH_0324|M.thermoresistible__bu EEPVEAP---------TLKHSPMLRVVFGHLTNPGRLKEVLAEHAAYADR
MSMEG_6226|M.smegmatis_MC2_155 NPQDDP----------PERSAELLRVFLLGELPPEQAREHLVMLASHSDG
Mvan_5461|M.vanbaalenii_PYR-1 ERQDDP----------PVRSAGLLRVFLLGLVPPEQARAHLHAMAEEADG
Mflv_1331|M.gilvum_PYR-GCK DFHDDP----------PQRSSALLRVFLLSAVPSEEAGKYMIAVAEHAER
Mb1209c|M.bovis_AF2122/97 EPLSPTRPGRGSALTDSSTRDIAVKLRGAGYGDVAALYTQVTALRAERVK
Rv1176c|M.tuberculosis_H37Rv EPLSPTRPGRGSALTDSSTRDIAVKLRGAGYGDVAALYTQVTALRAERVK
MAV_0135|M.avium_104 TAPRPS----------VIRDELLIKVQAADAGDMRAVRDAIRERRDWATA
. ::: :
MMAR_1544|M.marinum_M -VRAEGLPLVAELL---ATGGPFPQRLHLNERLVSFYSDFNNLLLRWCEE
MUL_2388|M.ulcerans_Agy99 -VRAEGLPFVAELL---ATGGPFPQRLHLNERLVSFYSDFNNLLLRWCEE
MAB_0082c|M.abscessus_ATCC_199 -MQRRAALDAAGAA---AQPAWAYSRVAL-KWAERYYAAERDLAMMMIED
TH_0324|M.thermoresistible__bu -MQREAATEARHTA---EQPAWSYARLAL-QWSERYYAAERDLALQLIKD
MSMEG_6226|M.smegmatis_MC2_155 EVERLRALEAAID-----WAGEP-GADVFSHAALDYGLRMHTMQAQWARD
Mvan_5461|M.vanbaalenii_PYR-1 EVRRLTELRESLTG---GRAADPTDESFFGFAALDYGLRLNAMQAEWARS
Mflv_1331|M.gilvum_PYR-GCK DLARYEALRDAVP-----WNQSD-DDDFYGRAALEFGLRQAAMTADWARW
Mb1209c|M.bovis_AF2122/97 SLDTYRGIEKRTFADPSALDGAA----LHQYLVLRGGIRAEESAIDWLDE
Rv1176c|M.tuberculosis_H37Rv SLDTYRGIEKRTFADPSALDGAA----LHQYLVLRGGIRAEESAIDWLDE
MAV_0135|M.avium_104 KLARYQRLRARLLDGRSEEDYLARAERIGPYLTLIRGISFEEDNIRWAEH
:
MMAR_1544|M.marinum_M TLADVETWQGTQDIGLTPSGRQRLVRAISEHP-------------
MUL_2388|M.ulcerans_Agy99 TLADVETWQGTQDIGLTPSGRQRLVRAISEDP-------------
MAB_0082c|M.abscessus_ATCC_199 LAEAAEEFDKAGPDGGTPRLVPGYWREVEKRVDAEKLSTQHDTAD
TH_0324|M.thermoresistible__bu LDVADRIFTEAE-QSGLEWPVREYWYEVERRIAAEKDRE------
MSMEG_6226|M.smegmatis_MC2_155 VIARMDTNR------------------------------------
Mvan_5461|M.vanbaalenii_PYR-1 VIDRLDT--------------------------------------
Mflv_1331|M.gilvum_PYR-GCK VADGIRARQK-----------------------------------
Mb1209c|M.bovis_AF2122/97 VAEALQEKR------------------------------------
Rv1176c|M.tuberculosis_H37Rv VAEALQEKR------------------------------------
MAV_0135|M.avium_104 ALAVIARRLPTTDADSDAGDSRLVGPATNG---------------