For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSVQLSMGIPSFSAEPPPSWEHLTDWAQLLEGCGFDRVLVSEHIAFGAHMDAYANPATGGTAGGRQPTG PDGHWLEPLTTLTYLAARTERIRLGTNILLAALRPAAVLAKTLATMDVLSGGRIDIGVGVGWQREEYEAA GADFDRRGAVLDQALEVCQRLWRENEVSFESVGLNFERIHQMPKPTQPGGVPIWVSGTVNRAVARRLARF GAGWIPWGPDARDVAAGIARMRAAVAHAGGDPNGFGVAGALRVKADADGRPDLADVAERAKTLRDAGVTE LRMTHWPLAPESRERDISAVVEAVRAATGG
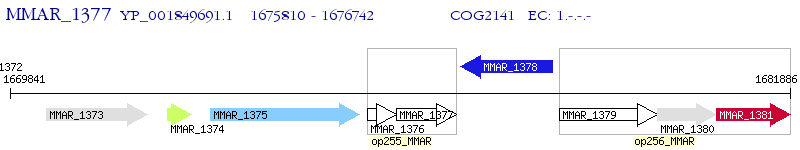
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1377 | - | - | 100% (310) | hypothetical protein MMAR_1377 |
| M. marinum M | MMAR_1586 | - | 3e-26 | 28.83% (274) | oxidoreductase |
| M. marinum M | MMAR_0393 | - | 7e-25 | 36.56% (186) | hypothetical protein MMAR_0393 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3106c | - | 1e-24 | 33.80% (213) | hypothetical protein Mb3106c |
| M. gilvum PYR-GCK | Mflv_1590 | - | 1e-28 | 30.42% (263) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3079c | - | 1e-24 | 33.80% (213) | hypothetical protein Rv3079c |
| M. leprae Br4923 | MLBr_00269 | - | 1e-12 | 28.08% (203) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0366 | - | 1e-35 | 36.03% (272) | hypothetical protein MAB_0366 |
| M. avium 104 | MAV_2935 | - | 2e-34 | 34.75% (282) | hypothetical protein MAV_2935 |
| M. smegmatis MC2 155 | MSMEG_2811 | - | 4e-31 | 37.55% (229) | hydride transferase 1 |
| M. thermoresistible (build 8) | TH_2945 | - | 1e-29 | 32.36% (275) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_2498 | - | 1e-177 | 97.11% (311) | hypothetical protein MUL_2498 |
| M. vanbaalenii PYR-1 | Mvan_5167 | - | 4e-29 | 31.56% (263) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3106c|M.bovis_AF2122/97 ----------MQFGVLTFVTDEGIG---PAELGAALEHRGFESLFLAEHT
Rv3079c|M.tuberculosis_H37Rv ----------MQFGVLTFVTDEGIG---PAELGAALEHRGFESLFLAEHT
MSMEG_2811|M.smegmatis_MC2_155 ----------MQISLHALGIGAGARREIIDAVAVAAERAGFTTLWCGEHV
MMAR_1377|M.marinum_M -----MTSVQLSMGIPSFSAEPPPSWEHLTDWAQLLEGCGFDRVLVSEHI
MUL_2498|M.ulcerans_Agy99 -----MTSVQLSMGIPSFSAEPPPSWEHLTDWAQLLEGCGFDRVLVSEHI
Mflv_1590|M.gilvum_PYR-GCK MQLVFPLPHMLRLPAMSQPWEGSVTGADQTRMAKCADAWGYDMIAVPEHF
Mvan_5167|M.vanbaalenii_PYR-1 MRLVFALPHMLRLPAMSQPWEASVTGADQTRMARCADQWGYDMIAVPEHF
TH_2945|M.thermoresistible__bu MKFTFALPHMMELDAMMQPWEMAVTGADQTRMARRAEELGFDMIAVPEHI
MAB_0366|M.abscessus_ATCC_1997 ---------MKIGVVAPVELGVTAEPDKILEFARAAERLGFSEISVVEHA
MAV_2935|M.avium_104 --------------MAPVADGVTADPDWMVAFARHLEACGFESIVAVEHT
MLBr_00269|M.leprae_Br4923 ---------MAELRLGYKASAEQFAPRELVELGVAAEAHGMDSATVSDHF
. : * :*
Mb3106c|M.bovis_AF2122/97 HIPVNTQSPYP-------GGGPIPEKYYRTLDPFVALAAAAATTQSLVLG
Rv3079c|M.tuberculosis_H37Rv HIPVNTQSPYP-------GGGPIPEKYYRTLDPFVALAAAAATTQSLVLG
MSMEG_2811|M.smegmatis_MC2_155 VMVDRPRSRYPYSA----DGRIAVAADADWLDPMIALSFAAAATSTIRIA
MMAR_1377|M.marinum_M AFGAHMDAYANPATGGTAGGRQPTGPDGHWLEPLTTLTYLAARTERIRLG
MUL_2498|M.ulcerans_Agy99 AFGAHMDAYANPATGGTAGGRQPTGPDGHWLEPLTTLTYLVARTERIRLG
Mflv_1590|M.gilvum_PYR-GCK VIPAEHVELSG----------------PHYLQSTVAQAYLAGATERIRLN
Mvan_5167|M.vanbaalenii_PYR-1 VIPAEHVELSG----------------PHYLQSTVAQAYLAGATERIRLN
TH_2945|M.thermoresistible__bu VMPREHVDLSG----------------GHWLHSTIAQAYFAGATERITLN
MAB_0366|M.abscessus_ATCC_1997 VVIGDTQSTYPYSPTG----QSHLPDDIDIPDPLELLSFVAAATTTLGLS
MAV_2935|M.avium_104 VLLTRYDSVYPYDSSG----RVGLAPDCPIPDPLDLLAFLAGHTTRLGLA
MLBr_00269|M.leprae_Br4923 QPWRHQG--------------------GHASFSLSWMTAVGERTNRILLG
. : * : :
Mb3106c|M.bovis_AF2122/97 TGIALIPERD-PIVTAKEVASLDLVSQGRFRFGVGVGWLREEVAN----H
Rv3079c|M.tuberculosis_H37Rv TGIALIPERD-PIVTAKEVASLDLVSQGRFRFGVGVGWLREEVAN----H
MSMEG_2811|M.smegmatis_MC2_155 TGVLLLPEHN-PVIMAKQAASVDQLSGGRLLLGVGIGWSREEFEA----L
MMAR_1377|M.marinum_M TNILLAALRP-AAVLAKTLATMDVLSGGRIDIGVGVGWQREEYEA----A
MUL_2498|M.ulcerans_Agy99 TNILLAALRP-AAMLAKTLATMDVLSGGRIDIGVGVGWQREEYEA----A
Mflv_1590|M.gilvum_PYR-GCK SCITVLPLQH-PIVLAKALATADWMSGGRMMVTFGVGWLKGEFDA----L
Mvan_5167|M.vanbaalenii_PYR-1 SCITVLPLQH-PVVLAKALATADWMSGGRMMVTFGVGWLQGEFEA----L
TH_2945|M.thermoresistible__bu SCVTILPLQH-PLVTAKALATADWLSGGRMMVTFGVGWLQREFEL----L
MAB_0366|M.abscessus_ATCC_1997 TGVLVLPDHH-PVVLAKRLATLDRLSRGRLRICAGVGWMREEIEA----C
MAV_2935|M.avium_104 TGVLVLPNHH-PVVLAKRVGTLDALSGGRLRLCVGVGWLQEELRA----C
MLBr_00269|M.leprae_Br4923 TSVLTPTFRYNPAVIGQAFATMGCLYPNRVFLGVGTGEALNEVATGYQGA
: : . : . : .: .: . : .*. . * * *
Mb3106c|M.bovis_AF2122/97 GVDPAVRGRVIDERLRAIIEIWTQ---EQAEFHGTYVDFDPIYCWPKPVT
Rv3079c|M.tuberculosis_H37Rv GVDPAVRGRVIDERLRAIIEIWTQ---EQAEFHGTYVDFDPIYCWPKPVT
MSMEG_2811|M.smegmatis_MC2_155 GVPFARRGARTAEYVEAMRTLWRQ---DVADFTGEFAAFDAVRLHPKPSA
MMAR_1377|M.marinum_M GADFDRRGAVLDQALEVCQRLWRE---NEVSFESVGLNFERIHQMPKPTQ
MUL_2498|M.ulcerans_Agy99 GVDFDRRGAVLDQTLEVCQRLWRE---NEVSFESAGLNFERIHQMPKPAQ
Mflv_1590|M.gilvum_PYR-GCK GVPFRERGRIADEYLAAIVELWTS---DSPSFDGRYVSFDEIAFEPKPVQ
Mvan_5167|M.vanbaalenii_PYR-1 GVPFRERGRMADEYLAVIKELWTS---DAPSFEGKYVSFREVAFEPKPVQ
TH_2945|M.thermoresistible__bu GVPFRKRGRIADEYLAAIIELWTQ---DEPSFAGQFVSFEDVVFEPKPIQ
MAB_0366|M.abscessus_ATCC_1997 GGDFDRRGRAADEAIAVMRALWST--DGPTSHDGEFYSFRNAYSYPKPVR
MAV_2935|M.avium_104 GADFDNRGRRADEQLAVLRALWAHRPDG-ASFHGEFFTFDNVMCYPKPVA
MLBr_00269|M.leprae_Br4923 WPEFKERFARLRESVRLMRELWRG---DRVDFDGDYYQLKGASIYDVP--
* : : :* .. . : *
Mb3106c|M.bovis_AF2122/97 KPY--PPLYVGGGP-ANFPRIARLNAGWIAISPSPQRLSGP------LQR
Rv3079c|M.tuberculosis_H37Rv KPY--PPLYVGGGP-ANFPRIARLNAGWIAISPSPQRLSGP------LQR
MSMEG_2811|M.smegmatis_MC2_155 GSI--PIIVGGNGD-RALARVASWGDGWYGFNLDGVAAVGER--MDMLRR
MMAR_1377|M.marinum_M PGG-VPIWVSGTVNRAVARRLARFGAGWIPWGPDARDVAAG---IARMRA
MUL_2498|M.ulcerans_Agy99 PGGGVPIWVSGTVNRAVARRLARFGAGWIPWGPDARDVAAG---IARMRA
Mflv_1590|M.gilvum_PYR-GCK RPH-LPIWIGGDAD-AALRRAAKYASGWWSFLTPPDKIAER---VEFIKS
Mvan_5167|M.vanbaalenii_PYR-1 KPH-LPIWIGGDAD-AALRRAAKYASGWWSFLTPPEKLAER---LDFIKS
TH_2945|M.thermoresistible__bu QPH-PPIWIGGDSD-AALRRAARFASGWFTFLTKPEDIAAR---VDIITS
MAB_0366|M.abscessus_ATCC_1997 PQG-VPLYIGGHSK-AAARRAGRLGSGFQPLGLVGDDLTAA---VGVMRA
MAV_2935|M.avium_104 AER-FPVHIGGHSR-AAARRAGRFGDGFQPLGVTGARLASL---IGLMRD
MLBr_00269|M.leprae_Br4923 -EGGVPIYIAAGGP-EVAKYAGRAGEGFVCTSGKGEELYTEKLIPAVLEG
* . . *: :
Mb3106c|M.bovis_AF2122/97 LRAMAG----------------GDVPVTVCQWG--EAAAKDLEGYRHLGV
Rv3079c|M.tuberculosis_H37Rv LRAMAG----------------GDVPVTVCQWG--EAAAKDLEGYRHLGV
MSMEG_2811|M.smegmatis_MC2_155 LCARAG----------------RDVDDLHCAVALQAPAVGDIPALVDLGV
MMAR_1377|M.marinum_M AVAHAG---GDPNGFG---VAGALRVKADADGRP--DLADVAERAKTLRD
MUL_2498|M.ulcerans_Agy99 AVAHAG---GDPNGFG---VAGALRVKADADGRP--DLADVAERTKTLRD
Mflv_1590|M.gilvum_PYR-GCK QDGYDGRPFDVVHGLGTNRVGEGHVAQKDPHARPGMSAAEIADKLNWLGE
Mvan_5167|M.vanbaalenii_PYR-1 QPDYDGRPFDVVHGLGTNRVGEGHVAQHDPDARPGMSAAEIVDRLSWLGA
TH_2945|M.thermoresistible__bu QPEYDGRPFDVMHGLGTNRVGEGHVPTGDRGAWPGMSAAELIDRIGALHE
MAB_0366|M.abscessus_ATCC_1997 AAVDAG---RDPRGLD---LVLGHGLHRVDQAALDQAAHAGAGRMLLSAS
MAV_2935|M.avium_104 EAVAAG---RDPAALE---VSLGHSVSKIDAERAARLADQGADRLVLAMP
MLBr_00269|M.leprae_Br4923 AAVAGRDADDIDKMIEIKMSYDPDPEQALSNIRFWAPLSLAAEQKHSIDD
Mb3106c|M.bovis_AF2122/97 ER-VLLELPTEPRDPTLRYLDKLQAELARLA-------------------
Rv3079c|M.tuberculosis_H37Rv ER-VLLELPTEPRDPTLRYLDKLQAELARLA-------------------
MSMEG_2811|M.smegmatis_MC2_155 DELVIVESPPQDPDVAADWVDALADRWITCI-------------------
MMAR_1377|M.marinum_M AGVTELRMTHWPLAPESRERDISAVVEAVRAATGG---------------
MUL_2498|M.ulcerans_Agy99 AGVTELRMTHWPLAPENRERDISAVVEAVRAATGG---------------
Mflv_1590|M.gilvum_PYR-GCK QGVTVSAVPIPKVSGVEEYLDYAQWVIEEIRPQVR---------------
Mvan_5167|M.vanbaalenii_PYR-1 QGVTVSAVPIPKVRGVEEYLDYAQWVIEEIKPQVP---------------
TH_2945|M.thermoresistible__bu HGVTYTSVPIPPVRGVEEYLDYSQWVMEEIAPNVP---------------
MAB_0366|M.abscessus_ATCC_1997 RRATTLEAVLEEMAGCATRLELG---GPR---------------------
MAV_2935|M.avium_104 P-TSDIEQAKDMLSACAQRLSLRPVSGP----------------------
MLBr_00269|M.leprae_Br4923 PIEMEKVADALPIEQVAKRWIVVSDPDEAVARVGQYVTWGLNHLVFHAPG
Mb3106c|M.bovis_AF2122/97 ----------------------
Rv3079c|M.tuberculosis_H37Rv ----------------------
MSMEG_2811|M.smegmatis_MC2_155 ----------------------
MMAR_1377|M.marinum_M ----------------------
MUL_2498|M.ulcerans_Agy99 ----------------------
Mflv_1590|M.gilvum_PYR-GCK ----------------------
Mvan_5167|M.vanbaalenii_PYR-1 ----------------------
TH_2945|M.thermoresistible__bu ----------------------
MAB_0366|M.abscessus_ATCC_1997 ----------------------
MAV_2935|M.avium_104 ----------------------
MLBr_00269|M.leprae_Br4923 HNQRRFLELFEKDLAPRLRRLG