For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLQELRMSEMRYRAVLEVLDGAPVTAVARRFGVSRQTVHAWLRRYAEEGAALNLEDRSSRPHRCPHQMPV EVEARVLTLRDAHPRWGPTRIVYELQRDGVPVVPGRSSVYRALVRNGRIDPAKRRRRRSDYKRWERGRPM ELWQMDVVGGLHLRDGIEVKVVTGIDDNSRFVVSAKVVARATARPVCAALLEALRRHGVPEQILTDNGKV FTGRFGPGGSSAEVLFDRVCVENGIGHLLTAPRSPTTTGKVERLHKTMRAEIFAEVDGVFDAIAELQAAI DRWVQYYNTARPHQSLGMVAPAARFALAAGPDLSVVEPVAAVPAEGQHPGALVDLRDAGVRRWVNRHGSI SVAGFRYRVPIVLAGEPVSVVVADNLVSIYHHDVLVASHVQHRKTDNTTGVVRQGRRTARPVTSGIVVTR VVDSNGDVSFAGTMYRAGRVWRGKQVEVSIVAESVQIACAGAIVRTHAIRHDRAKEHGAFATPHGRPRKP RQDAPEGPGVKATPLRGRPEGRALTPAP
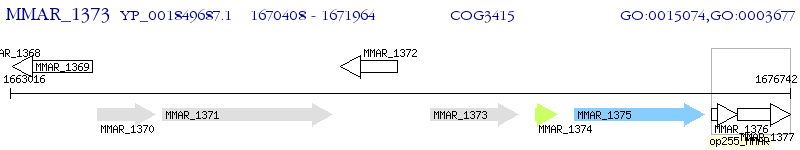
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1373 | - | - | 100% (518) | transposase for ISMyma05 |
| M. marinum M | MMAR_4106 | - | 0.0 | 99.42% (518) | transposase for ISMyma05 |
| M. marinum M | MMAR_4073 | - | 3e-20 | 30.89% (314) | transposase for ISMyma04 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2836 | - | 1e-09 | 26.80% (194) | transposase |
| M. gilvum PYR-GCK | Mflv_4763 | - | 4e-10 | 26.86% (242) | integrase catalytic subunit |
| M. tuberculosis H37Rv | Rv2812 | - | 1e-19 | 24.73% (372) | transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3347 | - | 5e-11 | 42.72% (103) | transposase |
| M. smegmatis MC2 155 | MSMEG_6600 | - | 2e-18 | 30.65% (323) | transposase |
| M. thermoresistible (build 8) | TH_3163 | - | 8e-33 | 30.50% (400) | PUTATIVE putative Integrase, catalytic region |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0573 | - | 1e-85 | 38.98% (531) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1373|M.marinum_M -----------------MLQELRMS------EMRYRAVLEVLDG-APVTA
Mvan_0573|M.vanbaalenii_PYR-1 -----------------MTQQLPNSPNDWVIEHRYRAVRQVLDG-VSKSQ
MAV_3347|M.avium_104 MKCELLKNAQLTPEVFVSHRNAPLS-----ETGRLRLARCVVEDGWSLRR
MSMEG_6600|M.smegmatis_MC2_155 ----------------MVHRNAPLS-----ETGRLRLARCVVDDGWSRRR
Mb2836|M.bovis_AF2122/97 --------------------------------------------------
Rv2812|M.tuberculosis_H37Rv ----MAVGDDEEKVRAERARAIGLFRYQLIWEAADAAHSTKQRGKMVREL
TH_3163|M.thermoresistible__bu -----------------MAQKVTAMD--------IRMAAAVAGEIDNVAE
Mflv_4763|M.gilvum_PYR-GCK ---------------MPASGDHRVDVAPLDCPVRRHESQRGQTAQGTRSR
MMAR_1373|M.marinum_M VARRFGVSRQTVHAWLRRYAEEGAALNLEDRSSRPHRCPHQMPVEVEARV
Mvan_0573|M.vanbaalenii_PYR-1 VAQECGASRQSVHSWVIRYEALGVAG-LADRSRRPLTSPNELSPAVVAMV
MAV_3347|M.avium_104 AAERFQVAVTTAQRWASRYRELGPAG-MADRSSRPLRSPNQTPTRTERRI
MSMEG_6600|M.smegmatis_MC2_155 AAERFQVSVTTVCRWVNRYLELGEAG-MADRSSRPHHSPNRTPTRTERRI
Mb2836|M.bovis_AF2122/97 --------------------------------------------------
Rv2812|M.tuberculosis_H37Rv ASREHTDPFGRRVRISRQTIDRWIRGWRAGGFDALVPNPRQCTPRTPAEV
TH_3163|M.thermoresistible__bu FCRRRKISRQTFYKFRRRFRDSGIEG-LQELSRRPLTSPGQTPVEVEDLI
Mflv_4763|M.gilvum_PYR-GCK ERPAQEAGRQPGPRHRHAQGDFGGKLLTPNRKRSAVIALRERFGVSERRA
MMAR_1373|M.marinum_M LTLR----DAHPRWGPTRIVYELQR--DGVPVVPGRSS-VYRALVRNGR-
Mvan_0573|M.vanbaalenii_PYR-1 CELR----RTYPRWGAQRIAHELAL--RGVDAPPSRSS-VYRILVRHGL-
MAV_3347|M.avium_104 IKVR----VLR-RWGPARIAYLLGLNVSTVHRVLTRYK-LAKLRWLDRA-
MSMEG_6600|M.smegmatis_MC2_155 IGVR----VTR-RWGPARIAYLLRLNVSTVHNVLRRYR-IARLRWLDRA-
Mb2836|M.bovis_AF2122/97 --------------------------------------------------
Rv2812|M.tuberculosis_H37Rv LELA----VALRRENPQRTAAAIRRILRTQLGWAPDER-TLQRNFHRLG-
TH_3163|M.thermoresistible__bu VLRRKQLIEQGRDHGAQSIVWSFERDGVTVPSVSTVWQRGWQILSRRGA-
Mflv_4763|M.gilvum_PYR-GCK CTVVGLHRSTMRLTPAPVTTEEAELRAWLRRFSTDRPRWGWRRAAKMARR
MMAR_1373|M.marinum_M IDPAKRRRRRSDYKRWERGRPMELWQMDVVGGLHLRDGIEVKVVTGIDDN
Mvan_0573|M.vanbaalenii_PYR-1 VAAQQQNHKR-KYRRWQRDAPMQLWQIDIMGGVFLVDGRECKVVTGIDDH
MAV_3347|M.avium_104 TGQVVRRMEAACCGDLVHVDVKKLGKIPAGGGWRMLGRTVGNRNAQADKT
MSMEG_6600|M.smegmatis_MC2_155 TGRVVRRMESAACGDLVHADVKKLGKIPAGGGWRMLGRTAGRKNSLADKS
Mb2836|M.bovis_AF2122/97 --------------------------------------------------
Rv2812|M.tuberculosis_H37Rv LTGATTGSAPAVFGRFEAEHPNALWTGDVLHGIRIDLRKTYLFAFLDDHS
TH_3163|M.thermoresistible__bu ITPQPQKRPKSATKRFVFSCPNECWQSDWTG-WVLADGSPAGIAGSLDDH
Mflv_4763|M.gilvum_PYR-GCK AGWKANNKRIRRLWREEGLRVPQRRRKKRLTGIGVAVGAMSPIRPNVIWA
MMAR_1373|M.marinum_M S----------RFVVSAKVVARATARPVCAALLEALRR------HGVPEQ
Mvan_0573|M.vanbaalenii_PYR-1 S----------RFVVMATVVADPGARAVCAAFTATMAI------YGVPSE
MAV_3347|M.avium_104 SGERSKHRHPLRGYHFLHTAIDAHSRLAYSELLTDERKETAAAFWLRAQA
MSMEG_6600|M.smegmatis_MC2_155 SGVKTRNGDPARGYHFLHSALDAHSRLVYSEMLADERKDTASEFWKRANS
Mb2836|M.bovis_AF2122/97 ------------------------MRLAAALRPALASR-------GVPNA
Rv2812|M.tuberculosis_H37Rv R----------LVPGYRWGHAEDTVRLAAALRPALASR-------GVPNA
TH_3163|M.thermoresistible__bu SR---------YVPALRACAGDADAQLVWETMLAGIAE------CGIPSM
Mflv_4763|M.gilvum_PYR-GCK MDFQFDTTADGRTLKMLNVIDEFTREALAIEVDRAINADG---VVDVLDR
. .
MMAR_1373|M.marinum_M ILTDNGKVFTGRFGPGGSSAEVLFDRVCVENGIGHLLTAPRSPTTTGKVE
Mvan_0573|M.vanbaalenii_PYR-1 VLTDNGKQFTGRFTKP-YPAEVLFERICRENGITTRLTKPRSPTTTGKIE
MAV_3347|M.avium_104 WFTTCGFTVRKVLTDNGSCYRSHAFRDALG-DIEHRRTRPYRPQTNGKVE
MSMEG_6600|M.smegmatis_MC2_155 WFFEQGVIVRTVLTDNGSCYRSHSFREALGDDIEHRRTRPYRPQTNGKVE
Mb2836|M.bovis_AF2122/97 VYVDNGS----------PYVDAWLLRACAKLGVRLVHSTPGRPQGRGKIE
Rv2812|M.tuberculosis_H37Rv VYVDNGS----------PYVDAWLLRACAKLGVRLVHSTPGRPQGRGKIE
TH_3163|M.thermoresistible__bu SLTDNGIVYTGR----FHAHQSAFETNLRALGVQTINATPFHPQTCGKIE
Mflv_4763|M.gilvum_PYR-GCK LALTYGAPHYVRFDNGPEFVANAVADWCRFNSAGSLFIDPGSPWQNAWIE
* . * * . :*
MMAR_1373|M.marinum_M RLHKTMRAEIFAEVDG--------VFDAIAELQAAIDRWVQYYNTARPHQ
Mvan_0573|M.vanbaalenii_PYR-1 RFHKTLRRELLDSAG---------PFASIEVAQEAIDAWVHGYNHSRPHQ
MAV_3347|M.avium_104 RFHRTLADEWAYARL----------YRSDAERCAEFPRWLHTYNHHRGHT
MSMEG_6600|M.smegmatis_MC2_155 RFHRTLADEWAYARL----------YTSDEQRCQEYPRWLHTYNHHRGHT
Mb2836|M.bovis_AF2122/97 RFFRTVREQFLVEITGEPDVVGRHYVADLAELNRLFTAWVETVYHRSVHS
Rv2812|M.tuberculosis_H37Rv RFFRTVREQFLVEITGEPDVVGRHYVADLAELNRLFTAWVETVYHRSVHS
TH_3163|M.thermoresistible__bu RFWQTLKKWLTARDP----------ATTIAELNALLEQFRTFYNHQRPHR
Mflv_4763|M.gilvum_PYR-GCK SFNGRLRDELLNLWR----------FDSLLEARVIIEDWRRDYNANRPHS
: : : *
MMAR_1373|M.marinum_M SLGMVAPAARFALAAGPDLSVVEPVAAVPAEGQHPGALVDLRDAGVRRWV
Mvan_0573|M.vanbaalenii_PYR-1 SLGMATPATMFRPAP---IEVVPPVP-VDTEPLTPDVVVPPRPRRHLPPA
MAV_3347|M.avium_104 ALGGQPPATRVPNLSGQYI-------------------------------
MSMEG_6600|M.smegmatis_MC2_155 ALGGQPPASRVPNLSGQYT-------------------------------
Mb2836|M.bovis_AF2122/97 ETG-QTPLARWSAGG-------------PIPLPAPETLTEAFLWEEHRRV
Rv2812|M.tuberculosis_H37Rv ETG-QTPLARWSAGG-------------PIPLPAPETLTEAFLWEEHRRV
TH_3163|M.thermoresistible__bu AHRGALPAEVFAAGD--------------KARPADRPVPAPVIVSRHTVG
Mflv_4763|M.gilvum_PYR-GCK AHGELTPAEFALQWT---------------TTHQPQVA------------
*
MMAR_1373|M.marinum_M NRHGSISVAG-------------------FRYRVPIVLAGEPVSVVVADN
Mvan_0573|M.vanbaalenii_PYR-1 TEHALIEVVHAVEWEAVLTPRARLLLPGDQQFKFTAALARREVTVWASDR
MAV_3347|M.avium_104 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
Mb2836|M.bovis_AF2122/97 TKTATVSLHG-------------------NRYEIDPALVGRKVELVFDPF
Rv2812|M.tuberculosis_H37Rv TKTATVSLHG-------------------NRYEIDPALVGRKVELVFDPF
TH_3163|M.thermoresistible__bu PTSGYVFVAP-------------------YKVNVGLRWAGHECDIIRDGQ
Mflv_4763|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1373|M.marinum_M LVSIYHHDVLVASHVQHRKTDN--TTGVVRQGRRTARPVTS-GIVVTRVV
Mvan_0573|M.vanbaalenii_PYR-1 SIHIVLDGTVIRTRPSRLSEHD--LRDLLRRGARIAGPEPARGAATIDIL
MAV_3347|M.avium_104 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
Mb2836|M.bovis_AF2122/97 DLTRIEVRLAGAPMGRAIPYHIGRHSHPKAKPETPTAPPKPSGIDYAQLI
Rv2812|M.tuberculosis_H37Rv DLTRIEVRLAGAPMRRAIPYHIGRHSHPKAKPETPTAPPKPSGIDYAQLI
TH_3163|M.thermoresistible__bu HITILSGTTLVRSFTADPTRNYQRGDKTTRTYRTREPKPPS---------
Mflv_4763|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1373|M.marinum_M DSNGDVSFAGTMYRAGRVWRGKQ---VEVSIVAESVQIACAGAIVRTHAI
Mvan_0573|M.vanbaalenii_PYR-1 TTSAVIEVARTVSRDGCIGLGGDKVLLESTLVGQRVTLRFEGALMHVVAN
MAV_3347|M.avium_104 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
Mb2836|M.bovis_AF2122/97 ETAHAAELARGVNYTALTGAADQIPGQLDLLTGQEAQPK-----------
Rv2812|M.tuberculosis_H37Rv ETAHAAELARGVNYTALTGAADQIPGQLDLLTGQEAQPK-----------
TH_3163|M.thermoresistible__bu --------------------------------------------------
Mflv_4763|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1373|M.marinum_M R------------HDRAKEHGAFAT-----PHGRPRKPRQDAPEGPGVKA
Mvan_0573|M.vanbaalenii_PYR-1 GRLVKTMPAPLPPDQRATLRGARPTSEPLPPPAPPNRAMRRVAANGTITI
MAV_3347|M.avium_104 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
Mb2836|M.bovis_AF2122/97 --------------------------------------------------
Rv2812|M.tuberculosis_H37Rv --------------------------------------------------
TH_3163|M.thermoresistible__bu --------------------------------------------------
Mflv_4763|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1373|M.marinum_M TPLRGRP----EGRALTPAP------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 AGQRLRVGRSYQGRTVAVAIDDTVFRVFIDGTEICTHARRTDTDITIFKA
MAV_3347|M.avium_104 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
Mb2836|M.bovis_AF2122/97 --------------------------------------------------
Rv2812|M.tuberculosis_H37Rv --------------------------------------------------
TH_3163|M.thermoresistible__bu --------------------------------------------------
Mflv_4763|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_1373|M.marinum_M -------
Mvan_0573|M.vanbaalenii_PYR-1 YPRRHGT
MAV_3347|M.avium_104 -------
MSMEG_6600|M.smegmatis_MC2_155 -------
Mb2836|M.bovis_AF2122/97 -------
Rv2812|M.tuberculosis_H37Rv -------
TH_3163|M.thermoresistible__bu -------
Mflv_4763|M.gilvum_PYR-GCK -------