For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVHRNAPLSETGRLRLARCVVDDGWSRRRAAERFQVSVTTVCRWVNRYLELGEAGMADRSSRPHHSPNRT PTRTERRIIGVRVTRRWGPARIAYLLRLNVSTVHNVLRRYRIARLRWLDRATGRVVRRMESAACGDLVHA DVKKLGKIPAGGGWRMLGRTAGRKNSLADKSSGVKTRNGDPARGYHFLHSALDAHSRLVYSEMLADERKD TASEFWKRANSWFFEQGVIVRTVLTDNGSCYRSHSFREALGDDIEHRRTRPYRPQTNGKVERFHRTLADE WAYARLYTSDEQRCQEYPRWLHTYNHHRGHTALGGQPPASRVPNLSGQYT
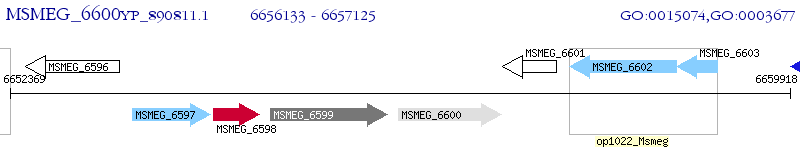
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6600 | - | - | 100% (330) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2619 | - | 1e-105 | 60.31% (325) | transposase for ISMyma06 |
| M. avium 104 | MAV_0752 | - | 1e-159 | 80.00% (330) | transposase |
| M. thermoresistible (build 8) | TH_2026 | - | 5e-67 | 59.15% (213) | transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0573 | - | 6e-21 | 30.41% (319) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6600|M.smegmatis_MC2_155 ----------------MVHRNAPLSETGRLRLARCVVDDGWSRRRAAERF
MAV_0752|M.avium_104 MKCELLKNAQLTPEVFVSHRNAPLSETGRLRLARCVVEDGWSLRRAAERF
MMAR_2619|M.marinum_M ----------------MSHANASLTPRGRLRLARCVVDDRWTYARAAERF
TH_2026|M.thermoresistible__bu --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 ------------MTQQLPNSPNDWVIEHRYRAVRQVLDG-VSKSQVAQEC
MSMEG_6600|M.smegmatis_MC2_155 QVSVTTVCRWVNRYLELGEAGMADRSSRPHHSPNRTPTRTERRIIGVRVT
MAV_0752|M.avium_104 QVAVTTAQRWASRYRELGPAGMADRSSRPLRSPNQTPTRTERRIIKVRVL
MMAR_2619|M.marinum_M QCSTATAKKWADRYRVGGPEAMVDRSSRPHRSPGRLPQRRERRIIKIRFT
TH_2026|M.thermoresistible__bu --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 GASRQSVHSWVIRYEALGVAGLADRSRRPLTSPNELSPAVVAMVCELRRT
MSMEG_6600|M.smegmatis_MC2_155 R-RWGPARIAYLLRLNVSTVHNVLRRYRIARLRWLDRATGRVVRR-----
MAV_0752|M.avium_104 R-RWGPARIAYLLGLNVSTVHRVLTRYKLAKLRWLDRATGQVVRR-----
MMAR_2619|M.marinum_M R-RWGPHRIAAHLRLARSTVEAVLRRYRMPLLRDLDQASGLPVRRPQPRR
TH_2026|M.thermoresistible__bu ----------------------------MPLLRHLDQNTGLPVRRPKPRR
Mvan_0573|M.vanbaalenii_PYR-1 YPRWGAQRIAHELALRGVDAPPSRSSVYRILVRHGLVAAQQQNHKRKYRR
:* : ::
MSMEG_6600|M.smegmatis_MC2_155 MESAACGDLVHADVKKLGKIPAGGGWRMLGRTAGRKNSLADKSSGVKTRN
MAV_0752|M.avium_104 MEAACCGDLVHVDVKKLGKIPAGGGWRMLGRTVGNRNAQADKTSGERSKH
MMAR_2619|M.marinum_M YEHPAPGDLVHLDVKKLGRIPDGGGHRKLGRHAGRRN-----RSG-----
TH_2026|M.thermoresistible__bu YEHPAPGDLVHVDVKKLGRIPDGGGHRKLGRQAGRRN-----RCG-----
Mvan_0573|M.vanbaalenii_PYR-1 WQRDAPMQLWQIDIMGGVFLVDGRECKVVTGIDDHSR-------------
: . :* : *: : * : : .. .
MSMEG_6600|M.smegmatis_MC2_155 GDPARGYHFLHSALDAHSRLVYSEMLADERKDTASEFWKRANSWFFEQGV
MAV_0752|M.avium_104 RHPLRGYHFLHTAIDAHSRLAYSELLTDERKETAAAFWLRAQAWFTTCGF
MMAR_2619|M.marinum_M ----VGYAFVHHAVDDHSRLAYSEILTDERKDTAAGFWLRANGFFVDHGI
TH_2026|M.thermoresistible__bu ----MGYTFLHHAVDDHSRLAYSEDLADERKETAAGFWKRASAFFADHGI
Mvan_0573|M.vanbaalenii_PYR-1 ------FVVMATVVADPGARAVCAAFT-----ATMAIYGVPSEVLTDNGK
: .: .: . . . :: :: :: .. : *
MSMEG_6600|M.smegmatis_MC2_155 IVRTVLTDNGSCYRSHSFREALGD--DIEHRRTRPYRPQTNGKVERFHRT
MAV_0752|M.avium_104 TVRKVLTDNGSCYRSHAFRDALG---DIEHRRTRPYRPQTNGKVERFHRT
MMAR_2619|M.marinum_M TVKRVLTDNGNCYRSKVFADSLGE--EIAHKRTRPYRPQTNGKVERFNRT
TH_2026|M.thermoresistible__bu TVKRVLTDNGSCYRSKNFAEALGP--DIAHKRTRPYRPQTNGKVERFNRT
Mvan_0573|M.vanbaalenii_PYR-1 QFTGRFTKP---YPAEVLFERICRENGITTRLTKPRSPTTTGKIERFHKT
. :*. * :. : : : * : *:* * *.**:***::*
MSMEG_6600|M.smegmatis_MC2_155 LADEWAYAR-LYTSDEQRCQEYPRWLHTYNHHRGHTALGGQPPAS--RVP
MAV_0752|M.avium_104 LADEWAYAR-LYRSDAERCAEFPRWLHTYNHHRGHTALGGQPPAT--RVP
MMAR_2619|M.marinum_M LASEWAYAQ-TYLSEVQRAATYQDWLHYYNHHRPHTGIGGKTPIERLRVH
TH_2026|M.thermoresistible__bu LTTEWAYAQ-TYRSEAERAGTYQHWLHHYNHHRPHTGIGGMTPIDRLRVH
Mvan_0573|M.vanbaalenii_PYR-1 LRRELLDSAGPFASIEVAQEAIDAWVHGYNHSRPHQSLGMATPATMFRPA
* * : : * *:* *** * * .:* .* *
MSMEG_6600|M.smegmatis_MC2_155 NLSGQYT-------------------------------------------
MAV_0752|M.avium_104 NLSGQYS-------------------------------------------
MMAR_2619|M.marinum_M NLPVKNT-------------------------------------------
TH_2026|M.thermoresistible__bu NLPVKNNQCPTSISRPPHCRGSKEAVSQFDALCANEPELAALRASVRDFL
Mvan_0573|M.vanbaalenii_PYR-1 PIEVVPPVPVDTEPLTPDVVVPPRPRRHLPPATEHALIEVVHAVEWEAVL
:
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0752|M.avium_104 --------------------------------------------------
MMAR_2619|M.marinum_M --------------------------------------------------
TH_2026|M.thermoresistible__bu KADRAEFGWQPDVDSWLGQWDEAFSARLGAAGFVGLTIPKKYGGAGLTHL
Mvan_0573|M.vanbaalenii_PYR-1 TPRARLLLPGDQQFKFTAALARREVTVWASDRSIHIVLDGTVIRTRPSRL
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0752|M.avium_104 --------------------------------------------------
MMAR_2619|M.marinum_M --------------------------------------------------
TH_2026|M.thermoresistible__bu HRYVVIEELLAQGAPVAAHWVADRQVVPSLLSYGSEDQRKRLLPRIAAGT
Mvan_0573|M.vanbaalenii_PYR-1 SEHDLRDLLRRGARIAGPEPARGAATIDILTTSAVIEVARTVSRDGCIG-
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0752|M.avium_104 --------------------------------------------------
MMAR_2619|M.marinum_M --------------------------------------------------
TH_2026|M.thermoresistible__bu FFSSIGMSEHGAGSDLAAVQTKAVSADGGWLLNGTKVWTSGAHKAHQVVV
Mvan_0573|M.vanbaalenii_PYR-1 -----------LGGDKVLLESTLVGQRVTLRFEGALMHVVANGRLVKTMP
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0752|M.avium_104 --------------------------------------------------
MMAR_2619|M.marinum_M --------------------------------------------------
TH_2026|M.thermoresistible__bu LARTSPPDRQMRHSGFSQFLVPCGTDGVRIEPILMMNGDHHFNEVSFEDV
Mvan_0573|M.vanbaalenii_PYR-1 APLPPDQRATLRGARPTSEPLPPPAPPNRAMRRVAANGTITIAGQRLRVG
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0752|M.avium_104 --------------------------------------------------
MMAR_2619|M.marinum_M --------------------------------------------------
TH_2026|M.thermoresistible__bu FVPESDVLGQIGQGWHQVTAELSFERSGPERILSTVPLIVAAIRVLGNTP
Mvan_0573|M.vanbaalenii_PYR-1 RSYQGRTVAVAIDDTVFRVFIDGTEICTHARRTDTDITIFKAYPRRHGT-
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0752|M.avium_104 --------------------------------------------------
MMAR_2619|M.marinum_M --------------------------------------------------
TH_2026|M.thermoresistible__bu TTDDTTAGILGDLLARLISLRQLSIWVARTLNESHAAANQAALVKDLGTQ
Mvan_0573|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0752|M.avium_104 --------------------------------------------------
MMAR_2619|M.marinum_M --------------------------------------------------
TH_2026|M.thermoresistible__bu FEQESTDVIIDVLNAVEATPELAALSATAMLHKPVFTLRGGTNEVLRGVV
Mvan_0573|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 -------
MAV_0752|M.avium_104 -------
MMAR_2619|M.marinum_M -------
TH_2026|M.thermoresistible__bu ARGMGLR
Mvan_0573|M.vanbaalenii_PYR-1 -------