For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SHRNARTTFHGRLLMVQRYRAGWFKARIAEAMGVSRKCVSTWIGRFEAEGMAGLLDRSSRPHTMPTRTSA VLENQIVAWRRRQRCGPEEIAAKLGVSARTVSRVLRRRGIPYLRDCDPMTGVVIRASKTTAVRYERARPG ELVHMDVKKLGRIPVGGGWRAHGRGRGRGGNRNRRHGSGFDYVHSLVDDHCRLAYSEILPDEKGATCAGF LRRAAAYFARHGIAGIEWVMTDNAWAYRYSLGAVCTEIGAQQIFIKPHCPWQNGKVERFNRTLQTEWAYR QVFTSNAQRNAALAPWLKHYNTQRRHGALGGLPPISRLQPTW*
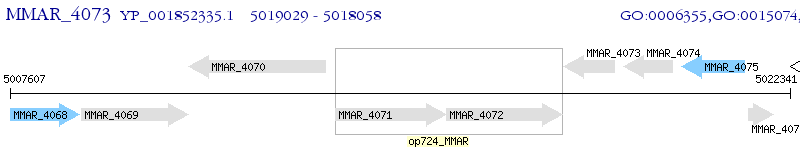
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4073 | - | - | 100% (323) | transposase for ISMyma04 |
| M. marinum M | MMAR_3857 | - | 0.0 | 100.00% (323) | transposase for ISMyma04 |
| M. marinum M | MMAR_3432 | - | 0.0 | 100.00% (323) | transposase for ISMyma04 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0603 | - | 7e-08 | 25.76% (132) | integrase catalytic subunit |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3347 | - | 9e-58 | 41.69% (331) | transposase |
| M. smegmatis MC2 155 | MSMEG_6600 | - | 7e-60 | 42.12% (330) | transposase |
| M. thermoresistible (build 8) | TH_2026 | - | 5e-49 | 49.52% (210) | transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0458 | - | 1e-20 | 30.33% (333) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_3347|M.avium_104 --------------------------------------------------
MSMEG_6600|M.smegmatis_MC2_155 --------------------------------------------------
TH_2026|M.thermoresistible__bu MPLLRHLDQNTGLPVRRPKPRRYEHPAPGDLVHVDVKKLGRIPDGGGHRK
MMAR_4073|M.marinum_M --------------------------------------------------
Mvan_0458|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0603|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3347|M.avium_104 -----------------------------MKCELLKNAQLTPEVFVSHRN
MSMEG_6600|M.smegmatis_MC2_155 ---------------------------------------------MVHRN
TH_2026|M.thermoresistible__bu LGRQAGRRNRCGMGYTFLHHAVDDHSRLAYSEDLADERKETAAGFWKRAS
MMAR_4073|M.marinum_M ---------------------------------------------MSHRN
Mvan_0458|M.vanbaalenii_PYR-1 --------------------------------------MVNPEVPESDSD
Mflv_0603|M.gilvum_PYR-GCK --------------------------------------------------
MAV_3347|M.avium_104 APLSETGRLRLARCVVEDGWSLRR-------------------------A
MSMEG_6600|M.smegmatis_MC2_155 APLSETGRLRLARCVVDDGWSRRR-------------------------A
TH_2026|M.thermoresistible__bu AFFADHG-ITVKRVLTDNGSCYRSKNFAEALGPDIAHKRTRPYRPQTNGK
MMAR_4073|M.marinum_M ARTTFHGRLLMVQRYR-AGWFKAR-------------------------I
Mvan_0458|M.vanbaalenii_PYR-1 VAARVTVEIRLRAVTEVNDGAPVG------------------------EV
Mflv_0603|M.gilvum_PYR-GCK -------------------------------------------------M
MAV_3347|M.avium_104 AERFQVAVTTAQRWASRYRELGPAG-------MADRSSRPLR--------
MSMEG_6600|M.smegmatis_MC2_155 AERFQVSVTTVCRWVNRYLELGEAG-------MADRSSRPHH--------
TH_2026|M.thermoresistible__bu VERFNRTLTTEWAYAQTYRSEAERAGTYQHWLHHYNHHRPHTGIGGMTPI
MMAR_4073|M.marinum_M AEAMGVSRKCVSTWIGRFEAEGMAG-------LLDRSSRPHT--------
Mvan_0458|M.vanbaalenii_PYR-1 AERYGVTRQTVTAWRKRYEAGGLEA-------LADQSRRPHS--------
Mflv_0603|M.gilvum_PYR-GCK SERLACKAVGLARSTYRRLPLGQTP---------DDPDAEMR--------
* .
MAV_3347|M.avium_104 ----------SPNQTPTRTER----------------------RIIKVRV
MSMEG_6600|M.smegmatis_MC2_155 ----------SPNRTPTRTER----------------------RIIGVRV
TH_2026|M.thermoresistible__bu DRLRVHNLPVKNNQCPTSISRPPHCRGSKEAVSQFDALCANEPELAALRA
MMAR_4073|M.marinum_M ----------MPTRTSAVLEN----------------------QIVAWRR
Mvan_0458|M.vanbaalenii_PYR-1 ----------SPGRISPDVEA----------------------LICEMRR
Mflv_0603|M.gilvum_PYR-GCK ----------AWLRSYATKHP--------------------------CHG
: . :
MAV_3347|M.avium_104 LRR-----------WGPAR--------IAYLLGLNVS--TVHRVLTRYKL
MSMEG_6600|M.smegmatis_MC2_155 TRR-----------WGPAR--------IAYLLRLNVS--TVHNVLRRYRI
TH_2026|M.thermoresistible__bu SVRDFLKADRAEFGWQPDVDSWLGQWDEAFSARLGAAGFVGLTIPKKYGG
MMAR_4073|M.marinum_M RQR-----------CGPEE----------IAAKLGVSARTVSRVLRRRGI
Mvan_0458|M.vanbaalenii_PYR-1 HHRR----------WGARR--------IAYELKLEIGKQAPSRSTTHRVL
Mflv_0603|M.gilvum_PYR-GCK FRR----------AWAALR----------YDEGREVNRKKIHRLWREEGL
* . .
MAV_3347|M.avium_104 AKLRWLDR--------------ATGQVVRRMEAACCGDLVHVDVKK--LG
MSMEG_6600|M.smegmatis_MC2_155 ARLRWLDR--------------ATGRVVRRMESAACGDLVHADVKK--LG
TH_2026|M.thermoresistible__bu AGLTHLHRYVVIEELLAQGAPVAAHWVADRQVVPSLLSYGSEDQRKRLLP
MMAR_4073|M.marinum_M PYLRDCDPMTG-------VVIRASKTTAVRYERARPGELVHMDVKK--LG
Mvan_0458|M.vanbaalenii_PYR-1 VRNGLVNSQEQ-----------QHKRVYKRWAREAPMHLWQLDIVG----
Mflv_0603|M.gilvum_PYR-GCK QVRVHSPRKR------------AGVSSVPPILADAPGVVWAIDFQF----
*
MAV_3347|M.avium_104 KIPAGGGWRMLGRTVG--NRNAQADKTSGERSKHRHPLRGYHFLHT-AID
MSMEG_6600|M.smegmatis_MC2_155 KIPAGGGWRMLGRTAG--RKNSLADKSSGVKTRNGDPARGYHFLHS-ALD
TH_2026|M.thermoresistible__bu RIAAGTFFSSIGMSEHGAGSDLAAVQTKAVSADGGWLLNGTKVWTSGAHK
MMAR_4073|M.marinum_M RIPVGGGWRAHGRGRG---------RGGNRNRRHG---SGFDYVHS-LVD
Mvan_0458|M.vanbaalenii_PYR-1 -----GVFLVNG--------------------------REHKMLTAIDDH
Mflv_0603|M.gilvum_PYR-GCK ------DSTVDG--------------------------KAIKIASMIDEH
* . .
MAV_3347|M.avium_104 AHSRLAYSELLTDERKETAAAFWLRAQAWFTTCGFTVRKVLTDNGSCYRS
MSMEG_6600|M.smegmatis_MC2_155 AHSRLVYSEMLADERKDTASEFWKRANSWFFEQGVIVRTVLTDNGSCYRS
TH_2026|M.thermoresistible__bu AHQVVVLARTSPPDRQMRHSGFSQFLVPCGTDGVRIEPILMMNGDHHFNE
MMAR_4073|M.marinum_M DHCRLAYSEILPDEKGATCAGFLRRAAAYFARHGIAGIEWVMTDNAWAYR
Mvan_0458|M.vanbaalenii_PYR-1 SRFVVAAAVLPVPSGPAVCDAFLAAIARWGAPFEVLTDNGKQFTGKFTRP
Mflv_0603|M.gilvum_PYR-GCK TRESLLN----LVERSITGEALVAELTKVFAAAGGPPKVLRMDNGPEMVS
: : . : .
MAV_3347|M.avium_104 HAFRDALG------------------DIEHRRTRP---------------
MSMEG_6600|M.smegmatis_MC2_155 HSFREALGD-----------------DIEHRRTRP---------------
TH_2026|M.thermoresistible__bu VSFEDVFVPESDVLGQIGQGWHQVTAELSFERSGPERILSTVPLIVAAIR
MMAR_4073|M.marinum_M YSLGAVCTEIG----------------AQQIFIKP---------------
Mvan_0458|M.vanbaalenii_PYR-1 LPAEVLFERICR------------DNGITARLTKR---------------
Mflv_0603|M.gilvum_PYR-GCK QALQRFCEN-----------------RTGMVYIPP---------------
.
MAV_3347|M.avium_104 ---YRPQTNGKVERFH------------------RTLADEWAYAR-LYRS
MSMEG_6600|M.smegmatis_MC2_155 ---YRPQTNGKVERFH------------------RTLADEWAYAR-LYTS
TH_2026|M.thermoresistible__bu VLGNTPTTDDTTAGILGDLLARLISLRQLSIWVARTLNESHAAAN-QAAL
MMAR_4073|M.marinum_M ---HCPWQNGKVERFN------------------RTLQTEWAYRQ-VFTS
Mvan_0458|M.vanbaalenii_PYR-1 ---RSPTTTGKVERFH------------------KTLRRELLDETGAFES
Mflv_0603|M.gilvum_PYR-GCK ---GCPWDNGYIESFN------------------NRLRRECLNRN-YWNT
* . : . * .
MAV_3347|M.avium_104 DAERCAEFPR-----WLHTYNHHRGHTALGGQPPATRVPN----------
MSMEG_6600|M.smegmatis_MC2_155 DEQRCQEYPR-----WLHTYNHHRGHTALGGQPPASRVPN----------
TH_2026|M.thermoresistible__bu VKDLGTQFEQESTDVIIDVLNAVEATPELAALSATAMLHKPVFTLRGGTN
MMAR_4073|M.marinum_M NAQRNAALAP-----WLKHYNTQRRHGALGGLPPISRLQP----------
Mvan_0458|M.vanbaalenii_PYR-1 IEAAQAAIDE-----WVHAYNTVRPHQSLDMATPASLFRSRVTDPELHPP
Mflv_0603|M.gilvum_PYR-GCK LFEARVVIGD-----FKHEHNHRHRHSALGYLTPAEYAAACR--------
. * . * ..
MAV_3347|M.avium_104 --LSGQYI------
MSMEG_6600|M.smegmatis_MC2_155 --LSGQYT------
TH_2026|M.thermoresistible__bu EVLRGVVARGMGLR
MMAR_4073|M.marinum_M -----TW-------
Mvan_0458|M.vanbaalenii_PYR-1 DLGHRRQIFGVLIR
Mflv_0603|M.gilvum_PYR-GCK --CTHTPGVCCMIR