For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTSMERKRQRNTTRVREAGAASRAQTRHLLLTAAAEEFARVGYVASTVSRIAEGAGVTVQTLYLAWGSKR ALLRGYLESTLAPDAAPSGQHFAAQLQPDSPAGTLAQVSALVCDAARRSAIAWRAYRDGSGVDSAIAAEW HQVQMLRRETMRALLVTIPDDALRLPRQDAVDTAWAITSPASHEIMVTHGGFTLQRFEAWLTTTLQNAIL RDPEGSG*
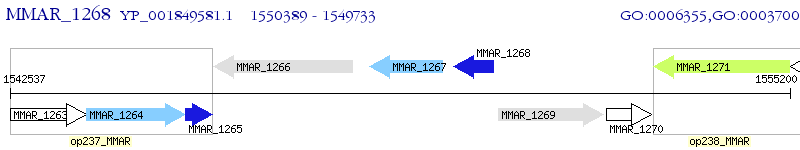
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1268 | - | - | 100% (218) | putative regulatory protein |
| M. marinum M | MMAR_3952 | - | 4e-07 | 28.49% (172) | hypothetical protein MMAR_3952 |
| M. marinum M | MMAR_0088 | - | 1e-06 | 28.12% (160) | transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4147 | - | 3e-07 | 44.29% (70) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4089c | - | 1e-06 | 32.48% (117) | TetR family transcriptional regulator |
| M. avium 104 | MAV_1977 | - | 1e-05 | 28.08% (146) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3976 | - | 1e-13 | 27.10% (214) | DNA-binding protein |
| M. thermoresistible (build 8) | TH_0688 | - | 3e-05 | 32.56% (86) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_2620 | - | 2e-97 | 99.44% (179) | hypothetical protein MUL_2620 |
| M. vanbaalenii PYR-1 | Mvan_2227 | - | 2e-05 | 40.00% (60) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1268|M.marinum_M MLTSMERKRQRNTTRVREAGAASRAQTRHLLLTAAAEEFARVGYVASTVS
MUL_2620|M.ulcerans_Agy99 ----MERKRQRNTTRVREAGAASRAQTRHLLLTAAAEEFARVGYVASTVS
MSMEG_3976|M.smegmatis_MC2_155 ----MKSEASRKHPYRSELRAQQAEATRQLVLDTATELFVERGYAATSID
MAV_1977|M.avium_104 ------------MTRRRAGEPRKGDLREAAILDAAEELLGAIGYDAMTIA
TH_0688|M.thermoresistible__bu -----MTTNSTSVDKAAGAGRPRDPRIDGAILRATADLLVEIGYSNLTMS
Mvan_2227|M.vanbaalenii_PYR-1 -----------MNEPASDAAAAEDTSTRHRILVATAEVLSRSGQTKLSLS
Mflv_4147|M.gilvum_PYR-GCK -MPVPQRNSPQKPQADVRNGRRPRGEARRLLLDAARELFARKDYRATTTR
MAB_4089c|M.abscessus_ATCC_199 ----------MSGRPRRISPIREGNSTRDEILDASAELFTTDGFAATTTR
:* :: : : . :
MMAR_1268|M.marinum_M RIAEGAGVTVQTLYLAWGSKRALLRGYLESTLAPDAAPS----GQHFAAQ
MUL_2620|M.ulcerans_Agy99 RIAEGAGVTVQTLYLAWGSKRALLRGYLESTLAPDAAPS----GQHFAAQ
MSMEG_3976|M.smegmatis_MC2_155 LIAQTAGVGRSTVFCAAGSKPWLLKTAYDRAVVGDDEQVPMVQRPQVRRL
MAV_1977|M.avium_104 DIAARAGITRGALYFYFGSKQEVITALFARTVAVLHEKS-------RAAT
TH_0688|M.thermoresistible__bu AVAERAGTTKTALYRRWSSKPELVYAATAPTDPVDTVES----------S
Mvan_2227|M.vanbaalenii_PYR-1 EVALQAGVSRPTLYRWFADKPALLEAFGIYEREMFETGI-----------
Mflv_4147|M.gilvum_PYR-GCK EIAEAAGVSEYLLFRHFGSKAGLFREALVVPFTSFLDQFRSTWQSVIPEE
MAB_4089c|M.abscessus_ATCC_199 RIAESVGIQQASLYYHFKTKDDILDALLAMTIDQPLHYA-------ALIA
:* .* :: * :.
MMAR_1268|M.marinum_M LQPDSPAGTLAQVSALVCDAARRSAIAWRAYRDGSGVDSAIAAEWHQVQM
MUL_2620|M.ulcerans_Agy99 LQPDSPAGTLAQVSALVCDAARRSAIAWRAYRDGSGVDSAIAAEWHQIQM
MSMEG_3976|M.smegmatis_MC2_155 FEMTDPTQIVEAYVEILGDAAQRVSRIYEVVRSAAGLDPEVHQLWTEIDG
MAV_1977|M.avium_104 EDAASGAKAIDTAMRRTEDLWREHGVVMRTAIDLAATVPQLDALWKQTAD
TH_0688|M.thermoresistible__bu DDVAGIVRALIERNRQITATPAARAAMPGLLVDMAADPRLHRGLLGRVGS
Mvan_2227|M.vanbaalenii_PYR-1 ----AKATAGLRGNDRVDAAMRFIVDYQQSYSGVRLVDIEPEVVIGRLAH
Mflv_4147|M.gilvum_PYR-GCK TDEEELSRQFVGQLYDVLVDHRGLLLTLVAADGLSAEDIEAAGIADIRRS
MAB_4089c|M.abscessus_ATCC_199 DRPEAPVVRLYALALGDAAQLAASRWNLGALYLLPDLRAERFSAFRRKRD
MMAR_1268|M.marinum_M LRRE---TMRALLVTIPDDALRLPRQDAVDTAWAITSPASH--------E
MUL_2620|M.ulcerans_Agy99 LRRE---TMRALLVTIPDDALRLPRQDAVDTAWAITSPASQPASQPATRS
MSMEG_3976|M.smegmatis_MC2_155 QRLEGADTVAALLKKKGGLKKGLTLAAARDLVWLYNDPGMH--------Y
MAV_1977|M.avium_104 IFVVAITAILEHMGVPAGKGPDKSAAIAEALCWMIERNFYQ--------A
TH_0688|M.thermoresistible__bu LVEA----LRRRLAEAAGRGEVQSGVDADRLTELIGGSVMFR------MV
Mvan_2227|M.vanbaalenii_PYR-1 IIPAMRAQLEKLLPGPNGAVKAATAIRVAVSHYIVRADDDDQ--------
Mflv_4147|M.gilvum_PYR-GCK LRVLG--QISAEGMQLRGLHSEQPDLPAHSTVATIVGMVAMR-----STF
MAB_4089c|M.abscessus_ATCC_199 QLRTHYQELAAAACRASMLRAGVERLPFRLVESVIMLRADGDS----IAP
:
MMAR_1268|M.marinum_M IMVTHGG--FTLQRFEAWLTTTLQNAILR---DPEGSG------------
MUL_2620|M.ulcerans_Agy99 WLPTAGSPCSALKPGSPPPCKTPSCAIRRAAGEPRRAAPPRRLVNRAEQR
MSMEG_3976|M.smegmatis_MC2_155 ALVEVRG--WSQRRYRAWLLDTFNHQLLGK--------------------
MAV_1977|M.avium_104 SRVSPARLTRARQTCTEIWLRVAS--------------------------
TH_0688|M.thermoresistible__bu LHPDEPITERWADEVAAILIRGVTKEG-----------------------
Mvan_2227|M.vanbaalenii_PYR-1 ---------FLAQLRHSVGIKPS---------------------------
Mflv_4147|M.gilvum_PYR-GCK FGENPPSREIIVDELVQAVLHGFLHRPDREPLSPKSVRPLQQ--------
MAB_4089c|M.abscessus_ATCC_199 EALADATLRVVGVTGDAGEIEGAARVLLGQLGWPVAIPKR----------
MMAR_1268|M.marinum_M ---------
MUL_2620|M.ulcerans_Agy99 PILMSHLTA
MSMEG_3976|M.smegmatis_MC2_155 ---------
MAV_1977|M.avium_104 ---------
TH_0688|M.thermoresistible__bu ---------
Mvan_2227|M.vanbaalenii_PYR-1 ---------
Mflv_4147|M.gilvum_PYR-GCK ---------
MAB_4089c|M.abscessus_ATCC_199 ---------