For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSAEQVAAKPGMARRRVGGIMVNPSRPGQRCAATSARRNERFEAFQHAISQTFVPLALCVDDVGTFRGQV RSASLGTVQLSDVRVGNDVLVRRTAKLINHSAPDYLKVAMPLNGKCVVTQDGHEETLAPRDFAVYDTTRP YSLAFSGAYRALVIMFPRDQVRLSSAELTRLTARRLNGRDGLGAPVSAFLAEMGNRLDDANTAGTLYLAD AVLDMLAASFAAQLSDETGGNAARGTRGLALRIRAFIESRLGDPGLDVSTIAAAHHVSVRYLQKLFESQG QTVTGWIRDRRIEHCRRDLEDHRLAEVPVSSIAARWGLVNASHFSRLFKAVHGSSPAEYRAQVSLRSPSA STDRG*
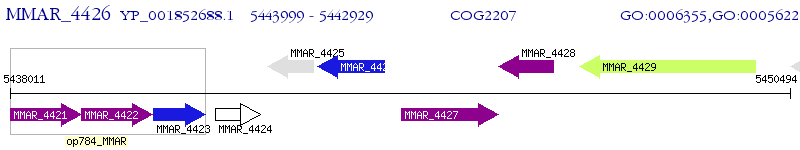
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4426 | - | - | 100% (356) | transcriptional regulatory protein |
| M. marinum M | MMAR_1063 | - | 2e-08 | 28.03% (157) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0731 | - | 8e-08 | 30.69% (101) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0715c | - | 9e-05 | 29.73% (111) | putative HTH-type transcriptional regulator AraC |
| M. avium 104 | MAV_4063 | - | 1e-06 | 31.71% (82) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_2260 | - | 9e-13 | 25.96% (312) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_1032 | - | 1e-09 | 27.17% (254) | putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_0821 | - | 2e-09 | 28.66% (157) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5518 | - | 2e-12 | 30.00% (160) | response regulator receiver protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_0821|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5518|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0731|M.gilvum_PYR-GCK --------------------------MHTSPSSTTSGDN---------LR
MAV_4063|M.avium_104 MGLTTMTVRDDGAPVGAPAEAARSDYGPVDPVLAVNGERRARPVAVEYKQ
MSMEG_2260|M.smegmatis_MC2_155 -------------MSRLTRLGFIDIRRTSDPVRRKVRSRGVPPALADHEI
TH_1032|M.thermoresistible__bu -------------MSRLTRLGFIDVRRTSNPVRRKVRSRGVPPSLVDNEI
MMAR_4426|M.marinum_M -------------MTSAEQVAAKPGMARRRVGGIMVNPSRPGQRCAATSA
MAB_0715c|M.abscessus_ATCC_199 ----------------------------------MELLSIPPRWGTLASE
MUL_0821|M.ulcerans_Agy99 ---------MDYLWRMAQAARLIQHRSGVPIYGY-RTDPDIPRVSVRRPG
Mvan_5518|M.vanbaalenii_PYR-1 ---------------MSETAVFAGRRDGTPVYAYPRRRPQAP-VSVLELG
Mflv_0731|M.gilvum_PYR-GCK FESDDLAETEDFLVRAYTKMSIAAGEGESP--SARIERWWLGSTTFDELE
MAV_4063|M.avium_104 FRGADAQSARRFFATAYDPGWRIAGVTNHC--VVSHRRSDTGSMTVDEVA
MSMEG_2260|M.smegmatis_MC2_155 FYTEDVREAARLVGRTLSPLTLTVDAADAGGFAATMHGVRLRNVSLLYLD
TH_1032|M.thermoresistible__bu FYTEDVAEAARLTGQVLAPLTLTVGAPDSAGFAATAHGVRLRNVSMLYLD
MMAR_4426|M.marinum_M RRNERFEAFQHAISQTFVPLALCVDDVGTFRGQVRSASLGTVQLSDVRVG
MAB_0715c|M.abscessus_ATCC_199 LAQQQGMDAAKMLIDAQISPRFLAGEPTMISPAQLRAVTREVLARTGDLS
MUL_0821|M.ulcerans_Agy99 PEDLGDRARHIHDF----PALWYVPAAGLVYIAAAGQVLEPTQFGSLQA-
Mvan_5518|M.vanbaalenii_PYR-1 PDAPPVRTTHIHEF----PVLVYQPATGSVDVVAAGLPVDPGRGGDRTG-
Mflv_0731|M.gilvum_PYR-GCK LDFHMSYDAAPLG-----RICLCRVHDGRIEENFIGEEPDVFAPGQVALL
MAV_4063|M.avium_104 IQGRMTLEIPAGD-----TVVVIQPRAG-----ALNVAGGPLATPDFPLL
MSMEG_2260|M.smegmatis_MC2_155 LHVAATLDIPESGR----YFAVHMPLNGSATVTHPAAEFDAGTTRSLVTS
TH_1032|M.thermoresistible__bu LHVPCALDIPRLGA----YFAVHMPLNGSAEVHHRGRQFQITPVRSLVTS
MMAR_4426|M.marinum_M NDVLVRRTAKLINHSAPDYLKVAMPLNGKCVVTQDGHEETLAPRDFAVYD
MAB_0715c|M.abscessus_ATCC_199 FG-SCQAPVPPEALRVLMFSLATSETAGAAFARWAAFRDAMPLVPSMTVA
*
MUL_0821|M.ulcerans_Agy99 -----GVAMFFDPAALGGDGGSPWPSWRTHPLLFPFLHGHSGGILRLEVP
Mvan_5518|M.vanbaalenii_PYR-1 -----AVAVFFDPAALGEGARSPWPTWRAHPLLSPFLHGHHDGILRLEVP
Mflv_0731|M.gilvum_PYR-GCK SPPELPYSGRVCGATFDLTMFDTALLDRVAAPANGRADQGIRLTGHRPVS
MAV_4063|M.avium_104 VAHGMACVLHCHGARFDVVTIAAEVLQKVAAEWHTALSQQTQFLNWRPRS
MSMEG_2260|M.smegmatis_MC2_155 PGDPVRMSFAHDSPQMLVRIEEEAMNAHLTRLLGRNLVGPLKFEPEFDLT
TH_1032|M.thermoresistible__bu PRGPLRIAFEHDSPQLVIRIEEAAMAAHLTRLLGRRLRRPLEFDPEFDMT
MMAR_4426|M.marinum_M TTR--PYSLAFSGAYRALVIMFPRDQVRLSSAELTRLTARR--LNGRDGL
MAB_0715c|M.abscessus_ATCC_199 RSGEFATVVLVSPSPKLPDVTFTEWTLALVIRVWSWLIMRAIHAERILLP
.
MUL_0821|M.ulcerans_Agy99 AARRSTWDGAIAAIEAELTTRQ---------QGYGQAVTAHLTLLLIELA
Mvan_5518|M.vanbaalenii_PYR-1 AARRPFWDAGVAALQAELTDRR---------DGYAQAALAHLTVLLIELA
Mflv_0731|M.gilvum_PYR-GCK PQAQQRLSDAIDYVRTVVRSDG-LEATPLVASTTASMLAAVVLHTLPSNA
MAV_4063|M.avium_104 RAAVRAWHRALDYAMLTLASPD-TARQPLIAAGMAGLLAGALLECYPSNL
MSMEG_2260|M.smegmatis_MC2_155 TEAATRWHAAVQLIHTEVYHAGSLVQRGCAIGPVEELAMSSLLYLQPSTY
TH_1032|M.thermoresistible__bu TEAAMRWHTAVQLIHTEVFHPGSLVQRGQGIGPVEELVMSSLLQLQPSTY
MMAR_4426|M.marinum_M GAPVSAFLAEMGNRLDDANTAG--------TLYLADAVLDMLAASFAAQL
MAB_0715c|M.abscessus_ATCC_199 QARPAGRSDRSNILKAPVEFGS-NSAAVILDAKLLDVPILRSEQEITSIL
MUL_0821|M.ulcerans_Agy99 RLARDVVGDLRRSGEPLLAEVFEVIDRRHAEP-LSLRDVAAEVGMTPGHL
Mvan_5518|M.vanbaalenii_PYR-1 RLSDDVVGDLRRSNEPLLAQVFDVIERRLGEP-LTLRDVARECGLTAGYL
Mflv_0731|M.gilvum_PYR-GCK SLEPT-ATDRLDAKPALLRRAIAFIEGNADSD-IALGDIAASVHVTPRAL
MAV_4063|M.avium_104 TEQDP-VSD--LALPETLKEAVSFIHRHAAED-IGINDVAAAVHLTPRAV
MSMEG_2260|M.smegmatis_MC2_155 YSDITRPTPAT-PRRAVIQQAVDYIEDHLAEK-ITMESLAEALHMSVRSI
TH_1032|M.thermoresistible__bu YDELLRPPPQPGQRRAVVRAAMNYIEEHLAER-ITVESVAKAVHVSVRSI
MMAR_4426|M.marinum_M SDETG--GNAARGTRGLALRIRAFIESRLGDPGLDVSTIAAAHHVSVRYL
MAB_0715c|M.abscessus_ATCC_199 DHPEQIWFDVGRYHRELPERVSRIVGSSIGQGIPTVSDIAAALNMAPSAL
: . : :* :: :
MUL_0821|M.ulcerans_Agy99 TTIVRRRTGRTVQEWIIERRMAQARELLV----ATDLAVGEVARRVGISD
Mvan_5518|M.vanbaalenii_PYR-1 TTVVRRRTGRTVGSWIAERRMAEARALLL----ATELTVGEVGRRVGMTD
Mflv_0731|M.gilvum_PYR-GCK QYMFRRHLEMTPMEYLRRVRMDHAHRDLLQADPAT-TTIQMVASRWGFGH
MAV_4063|M.avium_104 QYLFRRQLDTTPTEYMRRVRLSRAHQELVAATTAS-STVTEIAQRWGFAH
MSMEG_2260|M.smegmatis_MC2_155 QQGFRTELGMSPTTFIRERRLERVREELTDAIPSDGVTVTAVAERWGFHH
TH_1032|M.thermoresistible__bu QQNFRDELGLTPMEFVRERRLQRVHEELTDAIPSDGVTVTAVAQRWGFQH
MMAR_4426|M.marinum_M QKLFE-SQGQTVTGWIRDRRIEHCRRDLEDHR-LAEVPVSSIAARWGLVN
MAB_0715c|M.abscessus_ATCC_199 QRSLRNEFGTS----IRRIRDAALRTEAIKSLEDGSESLNDLSVRLGFSE
.. : : * : .: :. * *: .
MUL_0821|M.ulcerans_Agy99 PGYFSRLFSRAHGTSPRQWRALPPRH---------
Mvan_5518|M.vanbaalenii_PYR-1 PGYFTRQFRSCHGESPRTWRARHG-----------
Mflv_0731|M.gilvum_PYR-GCK TGRFASMYRDAYGRNPSDTLRN-------------
MAV_4063|M.avium_104 TGRFAVLYRQTYGQSPHTTLRQQTAV---------
MSMEG_2260|M.smegmatis_MC2_155 LGSFAVEYRKRWGETPSETLRR-------------
TH_1032|M.thermoresistible__bu LGSFAVEYRKRWGETPSETLRR-------------
MMAR_4426|M.marinum_M ASHFSRLFKAVHGSSPAEYRAQVSLRSPSASTDRG
MAB_0715c|M.abscessus_ATCC_199 LSAFTRAFRRWTGASPAQYRNGTRSVK--------
. *: : * .*