For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RSARGPHSGEPGRSGEPHSGRLGRSGEPHSGRLGRSGEPHSGRLGRSGEPHSVVVLALPGTIGFDLATAV EVFRRVYLADGSRPYLVRVCGTEPVVSAGPFGIATDLGLEAVSEAETIVVPARDNPTAPLPEGVLDALRG AYERGTRIASICAGAFTLAEAGILDGKRATTHWLAADLFRERYPAVHLDADVLYVDEGQVLTSAGAAAGL DLCLHIVARDHGAAVAAEAARVAVAPLHRDGGQAQYVIRNRRRAEVSLGEILAWIEHNAHRELSLEDLAT EASTSVRSLNRRFRAETGQTPMQWLTGVRVRHAQQLLERSADSVEWIGREVGFGSPANFREQFRRLTGVS PLAYRNTFRAQSA*
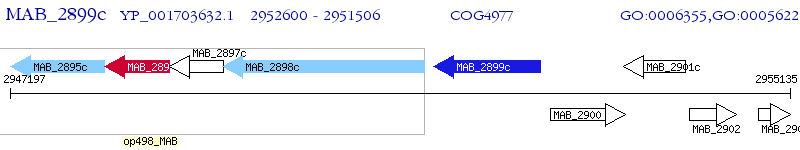
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2899c | - | - | 100% (364) | AraC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2445 | - | 5e-65 | 45.37% (313) | AraC family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0557 | - | 3e-56 | 37.62% (311) | AraC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0053 | - | 2e-10 | 29.29% (140) | hypothetical protein Mb0053 |
| M. gilvum PYR-GCK | Mflv_1693 | - | 2e-60 | 41.07% (319) | AraC family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0052 | - | 2e-10 | 29.29% (140) | hypothetical protein Rv0052 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2855 | - | 2e-40 | 33.44% (317) | transcriptional regulatory protein |
| M. avium 104 | MAV_4981 | - | 4e-44 | 34.78% (322) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_6839 | - | 1e-117 | 65.71% (315) | transcriptional regulator, AraC family protein |
| M. thermoresistible (build 8) | TH_3120 | - | 4e-54 | 39.49% (314) | PUTATIVE transcriptional regulator, AraC family with |
| M. ulcerans Agy99 | MUL_2902 | - | 2e-38 | 32.81% (317) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5349 | - | 1e-53 | 38.92% (316) | AraC family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2899c|M.abscessus_ATCC_199 MRSARGPHSGEPGRSGEPHSGRLGRSGEPHSGRLGRSGEPHSGRLGRSGE
MSMEG_6839|M.smegmatis_MC2_155 MR------------------------------------------------
TH_3120|M.thermoresistible__bu --------------------------------------------------
Mvan_5349|M.vanbaalenii_PYR-1 ------------------------------------------------MA
Mflv_1693|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_2855|M.marinum_M -------------------------------------------------M
MUL_2902|M.ulcerans_Agy99 -------------------------------------------------M
MAV_4981|M.avium_104 -------------------------------------------MADSSAT
Mb0053|M.bovis_AF2122/97 --------------------------------------------------
Rv0052|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2899c|M.abscessus_ATCC_199 PHSVVVLALPGTIGFDLATAVEVFRR----VYLADGSRPYLVRVCG---T
MSMEG_6839|M.smegmatis_MC2_155 --RVAVLAVPDVVAFDLSIPIEVFGR----VRLADGMNGYRVQVCG---T
TH_3120|M.thermoresistible__bu ---VAALVLDGLAIFEFGVICEVFGV----DRTDDGVPPVDFTVCGPRAG
Mvan_5349|M.vanbaalenii_PYR-1 IRSVSALVLDNLAMFEFGVICEVFGI----DRSADGVPNFDFKVCGPEAG
Mflv_1693|M.gilvum_PYR-GCK MHTVAILAYDGMSGFESGLAAEIFGMTELSERFSAGLTRPWYTVKLCSES
MMAR_2855|M.marinum_M ARKVVIVGFPGVQPLDVVGPFEVFAG-----ASLLSEGGYDVALASID-E
MUL_2902|M.ulcerans_Agy99 ARKVVTVGFPGVQPLDVVGPFEVFAG-----ASLLSEGGYDVALASID-E
MAV_4981|M.avium_104 PRVVVIVVFDDVTMLDVAGAGEVFAE-----ANRFG-ADYLLKIASVD-G
Mb0053|M.bovis_AF2122/97 ------------------------------------MPSFDVVFVGHRRG
Rv0052|M.tuberculosis_H37Rv ------------------------------------MPSFDVVFVGHRRG
.
MAB_2899c|M.abscessus_ATCC_199 EPVVSAGPFGIATDLGLEAVSEAETIVVPARDNP-TAPLPEGVLDALRGA
MSMEG_6839|M.smegmatis_MC2_155 EPTVSAGPLRIATDHGLEALADADTIIVPGRENL-PTQTPAPVVTALRDA
TH_3120|M.thermoresistible__bu QPVRTSFGATLTPDSGLDGLAGADLVAIPAIR---QDDYLPEALDAVRAA
Mvan_5349|M.vanbaalenii_PYR-1 SPLRTSVGATIIPDHGLDHLVGADLVAIPAIG---GSDYLPEALDAVRRA
Mflv_1693|M.gilvum_PYR-GCK AELRLLGGATVRTAYDLSDLAEADTVVIPSVRDI-GDPVSSELVDAIKTA
MMAR_2855|M.marinum_M RPVATGIGLEFMTTRLPDPSAPVDTVVLPGGGGVDAARDNVALMDWVKAA
MUL_2902|M.ulcerans_Agy99 RPVATGIGLEFMTTRLPDPSAPVDTVVLPGGGGVDAARDNVALMDWVKAA
MAV_4981|M.avium_104 RDVTTSIGTRLGVTDAVSSIESADTVLVAGSDNLPRRPIDPELVEAVKSV
Mb0053|M.bovis_AF2122/97 EVRSDNAMLGLLCDAAFDELTRPDVVIFPGGIGTRTLIHDQTVLDWVREA
Rv0052|M.tuberculosis_H37Rv EVRSDNAMLGLLCDAAFDELTRPDVVIFPGGIGTRTLIHDQTVLDWVREA
. . : : ... : :: .
MAB_2899c|M.abscessus_ATCC_199 YERGTRIASICAGAFTLAEAGILDGKRATTHWLAADLFRERYPAVHLDAD
MSMEG_6839|M.smegmatis_MC2_155 YASGATIASICTGAFVLADTGLLDGRRATTHWAAADLFRANFPAVDLDPD
TH_3120|M.thermoresistible__bu ADAGAIILTVCSGAFLAGAAGLLDGRPCTTHWGLTEELARRYPTARVDRN
Mvan_5349|M.vanbaalenii_PYR-1 AESGSIILTVCSGAFVAGAAGLLDGRPCTTHWMHADELARMYPTAKVDRN
Mflv_1693|M.gilvum_PYR-GCK DARGARLVSICSGAFALAAAGVLDGHKATTHWIYTDELRETYPGIDIDPN
MMAR_2855|M.marinum_M AGTARRVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAREFPAVDVDPD
MUL_2902|M.ulcerans_Agy99 AGTARRVVTVCTGTFLAAEAGLLDGCRATTHWAFADRLAREFPAVDVDPD
MAV_4981|M.avium_104 ADRTRRLASICTGSFILGQAGLLDGRRATTHWHDVRLFARAFPEITVEPD
Mb0053|M.bovis_AF2122/97 HRHTLLTTSVCTGGLVLAAAGLLNGLTATTHWRVQDLFNSLGARYVPQRV
Rv0052|M.tuberculosis_H37Rv HRHTLLTTSVCTGGLVLAAAGLLNGLTATTHWRVQDLFNSLGARYVPQRV
::*:* : . :*:*:* .**** : . :
MAB_2899c|M.abscessus_ATCC_199 VLYVDEG-QVLTSAGAAAGLDLCLHIVARDHGAAVAAEAARVAVAPLHRD
MSMEG_6839|M.smegmatis_MC2_155 VLYVDAG-RILTSAGASAGVDLCLHMVQRDHGAAAAANAAKMAVAPLHRS
TH_3120|M.thermoresistible__bu VLYVDDG-DLITSAGTAAGIDACLHLVRRELGSEVTNTIARHMVVPPQRD
Mvan_5349|M.vanbaalenii_PYR-1 VLFVDDG-NLITSAGTAAGIDACLHLVRRELGSEVTNKIARRMVVPPQRD
Mflv_1693|M.gilvum_PYR-GCK PLYVDNG-RVLTSAGCAAGLDLCLHIVRTDHGVRVANDVARRLVISPHRA
MMAR_2855|M.marinum_M PIFIRSSPKVWTAAGVTAGIDLALALVEGDHGTEIAQTVARWLVLYLRRP
MUL_2902|M.ulcerans_Agy99 PIFIRSSPKVWTAAGVTAGIDLALALVEGDHGTEIAQTVARWLVLYLRRP
MAV_4981|M.avium_104 ALFVCDG-DVFTSAGISSGIDLALALVEMDYGTELVRAVARWLVVYLKRA
Mb0053|M.bovis_AF2122/97 VEHLPER--VITAAGVSSGIDMGLRLVELLVSREAAEASQ----------
Rv0052|M.tuberculosis_H37Rv VEHLPER--VITAAGVSSGIDMGLRLVELLVSREAAEASQ----------
.: : *:** ::*:* * :* . .
MAB_2899c|M.abscessus_ATCC_199 GGQAQYVIRNRRRAEVS-----LGEILAWIEHNAHRELSLEDLATEASTS
MSMEG_6839|M.smegmatis_MC2_155 GGQAQFIIRNQPPASVIGEKTHLSEVLVWIEQNAHRELTLSDIADHAATS
TH_3120|M.thermoresistible__bu GGQRQFIDQPIPVRHSEG----FAPHLDWILSNLDKTHTVASLARRANMS
Mvan_5349|M.vanbaalenii_PYR-1 GGQRQYIDQPIPVKCSER----FAPHLDWILANLDKPHTVTTLARRAHMS
Mflv_1693|M.gilvum_PYR-GCK GGQAQYIETPVPEPTEDGR---IAAGMAWALEHLDETITLDELAARSAMS
MMAR_2855|M.marinum_M GGQTQFAAPVWMPRAKRPS---IRDVQEAIEAQPGGPHSIEELAHRAAMS
MUL_2902|M.ulcerans_Agy99 CGQTQFAAPVWMPRAKRPS---IRDVQEAIEAQPGGPHSIEELAHRAAMS
MAV_4981|M.avium_104 GGQSQFSALVEAEPPPRSP---LRKVTAAISADPAADHSVKSLSARASLS
Mb0053|M.bovis_AF2122/97 ---------------------------LMIEYDPQPPVDAGSLAKASPAT
Rv0052|M.tuberculosis_H37Rv ---------------------------LMIEYDPQPPVDAGSLAKASPAT
. :: : :
MAB_2899c|M.abscessus_ATCC_199 VRSLNRRFRAETGQTPMQWLTGVRVRHAQQLLERSADSVEWIGREVGFGS
MSMEG_6839|M.smegmatis_MC2_155 IRTLNRRFQAETGQTPMQWVTGVRIRHAQELLERTSLSVERIGRDVGFTS
TH_3120|M.thermoresistible__bu PRTFARRFVEETGRTPMQWVTDQRVLYARRLLEETDLDIHRIAARCGFGS
Mvan_5349|M.vanbaalenii_PYR-1 GRTFARRFVEETGRTPMQWVTDQRVLFARRMLEETNLDIDRIAEQSGFGT
Mflv_1693|M.gilvum_PYR-GCK RRSYLRQFAKATGTTPIRWLIEQRIQASLALLESSDHSIDQIAARVGFES
MMAR_2855|M.marinum_M PRHFTRVFTGEVGEAPRQYVERVRTEAARRQLEETEDTVVAIAARCGFGT
MUL_2902|M.ulcerans_Agy99 PRHFTRVFTGEVGEAPGRYVERVRTEAARRQLEETEDTVVAIAARCGFGT
MAV_4981|M.avium_104 TRQLTRLFQSELGTTPARYVEMVRIDAARAALDAGR-SVAESARLAGFGS
Mb0053|M.bovis_AF2122/97 HRLALEFYQHRL--------------------------------------
Rv0052|M.tuberculosis_H37Rv HRLALEFYQHRL--------------------------------------
* . :
MAB_2899c|M.abscessus_ATCC_199 PANFREQFRRLTGVSPLAYRNTFRAQSA-------------
MSMEG_6839|M.smegmatis_MC2_155 AANFREQFRRLSGVAPQVYRNTFRERTQDIPHTTSTALASA
TH_3120|M.thermoresistible__bu TTLLRHHFRRVIGVTPSDYRRRFACGSTTSGSTESGAEVTD
Mvan_5349|M.vanbaalenii_PYR-1 ATLLRHHFRRIIGVTPSDYRRRFN--STASQDVAVIA----
Mflv_1693|M.gilvum_PYR-GCK TATFRHHFVRQMHTTPSDYR------SAFTGS---------
MMAR_2855|M.marinum_M AETMRRNFIRRVGISPDQYRKAFA-----------------
MUL_2902|M.ulcerans_Agy99 AETMRRNFIRRVGISPDQYRKAFA-----------------
MAV_4981|M.avium_104 AETLRRVFVEHLGLSPKAYRDRFRTAARG------------
Mb0053|M.bovis_AF2122/97 -----------------------------------------
Rv0052|M.tuberculosis_H37Rv -----------------------------------------