For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNFSVLPPEVNSALIFTGPGAEPMRTAATAWGGMAVELESAADSFASVTSGLVGGAWQGASSVAMGTAAA SYLGWLNAAAAHAEQAAAQAEALVAEFAAAQSAMVQPFLLAANRSSLVSLVLSNLFGQNAPAIAAVEAEY EQMWALDVSTMATYHAGASAAAASLTPLAQPLQNLSDLPGRVIAAFGNGAPATTVTAAVTALRLPADAST VNFGLANIGSGNVGNANNGIGNLGSGNLGNFNLGSGNLGSANAGWANLGSNNIGGANSGGNNLGWGNLGG LNTGFANAGSGNFGFANTGNNNIGIGLTGDNQVGIGGLNSGVGNFGLFNSGTGNVGLFNSGTGNFGIGNS GAYGTGLFNSGQANTGLLNAGSFNTGLLDVGNANTGSLNTGSFNMGSFNPGPSNTGALNTGTANTGWFNT GSLNTGAFNIGDMNNGLFNTGEMNNGIFYRGVGQGNFYFSITTPDLTLPPLEIPGITIPNLSLPPITVPA LTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFD LPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPA NVVVGGFDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALT IPAATTPANVVVGGFDLPQLGIPSLTIPPLTLAEGTTVGAFSLPELGIPSLTIPPLTLAEGTTVGAFSLP ELGIPSLTIPPLTLAEGTTVGAFSLPQLGIPSLTIPSLTVPAGTTVGGFDLPALQTRSITVGGIDAGGFR TPAISTVSSTGYLELPTVTIPGVRVNNFVAFIPNNIEFLQTGQPGVIPQIGGLEHQAYISTGDIVIQGFS ISGIGFSLPQIDVGAFTLPELTIPPIGIPPTIVGAFTLPEITTPVITTPPLVVDPVSVAGFTLPEITTPE FTTPPLVVDPVSVAGFTLPEITTPEFTTPPLVVDPVSVAGFTLPEITTPEFTTPPLVVDPVSVAGFTLPQ ISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAG FTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDP VSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPQFTTAP ITIPPIGLGSFTTPPLTIPSIHLPGTTIGEFAFPAGPGYLNSSATPSSGFFNSGAGGNSGLGNNGSGLSG WFNTNPVGLLGGSGYQNYGGLSSGFSNLGSGVSGFANTGVRPFSLTGLISGFANIGSNLSGFFGSNVP
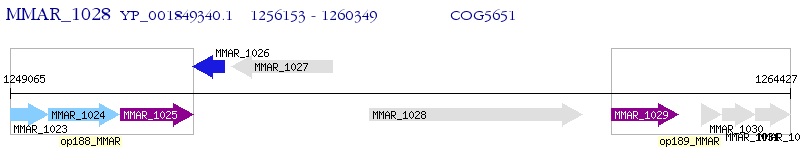
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_1028 | - | - | 100% (1398) | PPE family protein |
| M. marinum M | MMAR_0932 | - | 0.0 | 59.06% (1175) | PPE family protein |
| M. marinum M | MMAR_2822 | - | 0.0 | 54.18% (980) | PPE family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1782c | PPE24 | 0.0 | 54.15% (999) | PPE family protein |
| M. gilvum PYR-GCK | Mflv_0325 | - | 4e-24 | 33.33% (213) | PPE protein |
| M. tuberculosis H37Rv | Rv1753c | PPE24 | 0.0 | 56.20% (1025) | PPE family protein |
| M. leprae Br4923 | MLBr_01182 | - | 1e-26 | 27.25% (334) | PPE-family protein |
| M. abscessus ATCC 19977 | MAB_0809c | - | 2e-24 | 35.09% (171) | PPE family protein |
| M. avium 104 | MAV_4704 | - | 6e-51 | 51.94% (206) | PPE family protein |
| M. smegmatis MC2 155 | MSMEG_0619 | - | 8e-25 | 39.16% (166) | PPE family protein |
| M. thermoresistible (build 8) | TH_3113 | - | 4e-29 | 39.81% (206) | PUTATIVE PPE protein |
| M. ulcerans Agy99 | MUL_0782 | - | 0.0 | 98.77% (488) | PPE family protein |
| M. vanbaalenii PYR-1 | Mvan_0415 | - | 3e-26 | 40.96% (166) | PPE protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0325|M.gilvum_PYR-GCK MTSPVASLAPVTSPVWMALPPEVHSTMLSSGPGPGSLLAAGVAWQSLSAE
Mvan_0415|M.vanbaalenii_PYR-1 ----------MSSPVWMALPPEVHSALLSSGPGPGSMLAAAATWQSLSAE
MSMEG_0619|M.smegmatis_MC2_155 ----------MTAPIWMALPPEVHSSLLSSGPGPGSLLAAAGAWQSLSAE
MAB_0809c|M.abscessus_ATCC_199 ---------MLAAPVWMASPPEVHSALLSSGPGPGPLLAAAAEWSSLGAH
TH_3113|M.thermoresistible__bu ----------------MALPPEVHSTLLRTGPGPGALSAAADAWRMLSDE
MMAR_1028|M.marinum_M ----------MN---FSVLPPEVNSALIFTGPGAEPMRTAATAWGGMAVE
MUL_0782|M.ulcerans_Agy99 ----------MN---FSVLPPEVNSALIFTGPGAEPMRTAATAWGGMAVE
Mb1782c|M.bovis_AF2122/97 ----------MN---FSVLPPEINSALIFAGAGPEPMAAAATAWDGLAME
Rv1753c|M.tuberculosis_H37Rv ----------MN---FSVLPPEINSALIFAGAGPEPMAAAATAWDGLAME
MAV_4704|M.avium_104 ----------MTNPHFAWLPPEVNSALIYSGPGPGPLLAAAAAWDGLAEE
MLBr_01182|M.leprae_Br4923 ------------MFDFAALSPETNSTRMYLGPGSSPILTAAAAWVVLAKE
.** :*: : *.*. .: :*. * :. .
Mflv_0325|M.gilvum_PYR-GCK YASAAAELTSILAGVQSGAWEGPSSQQYVAAHTPYLAWLAQQSAVSAATA
Mvan_0415|M.vanbaalenii_PYR-1 YAAAAAELSSILADVQAGAWEGPSSEQYVAAHTPYLAWLAQQSAAGAAAA
MSMEG_0619|M.smegmatis_MC2_155 YAAAAAELTSVLSAVQAGSWEGPSSEQYVAAHAPYLQWLAQQSANSAAAA
MAB_0809c|M.abscessus_ATCC_199 YSATAQELRAILAATHASAWHGPSAETYVAAHTPYLEWLNQGALESNARA
TH_3113|M.thermoresistible__bu YRAAAVELAAVVSQTCATAWEGVGADAYAAAHGPYLFWLTDTAAAAAAAG
MMAR_1028|M.marinum_M LESAADSFASVTSGLVGGAWQGASSVAMGTAAASYLGWLNAAAAHAEQAA
MUL_0782|M.ulcerans_Agy99 LESAADSFASVTSGLVGGAWQGASSVAMGTAAASYLGWLNAAAAHAEQAA
Mb1782c|M.bovis_AF2122/97 LASAAASFGSVTSGLVGGAWQGASSSAMAAAAAPYAAWLAAAAVQAEQTA
Rv1753c|M.tuberculosis_H37Rv LASAAASFGSVTSGLVGGAWQGASSSAMAAAAAPYAAWLAAAAVQAEQTA
MAV_4704|M.avium_104 LASSAQSFSSVTSDLASGSWQGASSAAMMTVANQYVSWLSAAAAQAEEVS
MLBr_01182|M.leprae_Br4923 LTAAAQGLQSAVEALLT-TFEGESAAALAERVTPYEKWLTQNAASAELTA
::* : : ::.* .: * ** : . .
Mflv_0325|M.gilvum_PYR-GCK VQHDTAAAAYTTALATMPTLPELALNHTTHAVLVGTNFLGINTIPIALNE
Mvan_0415|M.vanbaalenii_PYR-1 AQHEAAAAAYSTALATMPTLPELALNHTTHAVLVATNFLGINTIPIAMTE
MSMEG_0619|M.smegmatis_MC2_155 VQHETAAAAYSTALATMPTMAELALNHTMHGVLVATNFFGINTIPIALNE
MAB_0809c|M.abscessus_ATCC_199 VQHGAAAGAYSTALAVMPTLAELAANHATHAALVATNFFGINTIPILATE
TH_3113|M.thermoresistible__bu AQLEAAAAAHASALAAMPTMAELTANRTTHAALVATNFFGVNTIPIAVNE
MMAR_1028|M.marinum_M AQAEALVAEFAAAQSAMVQPFLLAANRSSLVSLVLSNLFGQNAPAIAAVE
MUL_0782|M.ulcerans_Agy99 AQAEALVAEFAAAQSAMVQPFLLAANRSSLVSLVLSNLFGQNAPAIEAVE
Mb1782c|M.bovis_AF2122/97 AQAAAMIAEFEAVKTAVVQPMLVAANRADLVSLVMSNLFGQNAPAIAAIE
Rv1753c|M.tuberculosis_H37Rv AQAAAMIAEFEAVKTAVVQPMLVAANRADLVSLVMSNLFGQNAPAIAAIE
MAV_4704|M.avium_104 HQASAIATAFEVALAATVQPAVVAANRALVQALAATNWLGQNTPAIADIE
MLBr_01182|M.leprae_Br4923 TQLTVAANAYETAFTMTVPPLMVFVNRAQACLLIMSNIFGQNSTAIAEKE
* . . . : : *:: * :* :* *: .* *
Mflv_0325|M.gilvum_PYR-GCK ADYVRMWIQAATTMATYQSVSGAALAATPTSAPAPSVLLPGVGEAGRAAA
Mvan_0415|M.vanbaalenii_PYR-1 ADYVRMWIQAATTMATYQAVSGAALAATPTATPAPFVLAPGVGEAGRAAA
MSMEG_0619|M.smegmatis_MC2_155 ADYVRMWIQAATTMSTYQAVSGAAVAAAPVSAPAPFLLKPGVGEAGSASA
MAB_0809c|M.abscessus_ATCC_199 ADYVRMWVQAATTMATYQTASEVALASAPPSTPPPSILHD----------
TH_3113|M.thermoresistible__bu ADYARMWIQAATTMATYEAVASAAVSATPRTVAPPPILGAGTGESATAAI
MMAR_1028|M.marinum_M AEYEQMWALDVSTMATYHAGASAAAASLTPLAQPLQNLSDLPGRVIAAFG
MUL_0782|M.ulcerans_Agy99 AEYEQMWALDVSTMATYHAGASAAAASLTPLAQPLQNLSELPGRVIAAFG
Mb1782c|M.bovis_AF2122/97 ATYEQMWAADVSAMSAYHAGASAIASALSPFSKPLQNLAGLP----AWLA
Rv1753c|M.tuberculosis_H37Rv ATYEQMWAADVSAMSAYHAGASAIASALSPFSKPLQNLAGLP----AWLA
MAV_4704|M.avium_104 AAYEQMWASDVAAMFGYHADASAAVAKLPPWNEVLQN-----------LG
MLBr_01182|M.leprae_Br4923 AEYTEMWIQDAAAMTSYQASVLEAVGATKAFTAPPLGVNEVG------LA
* * .** .::* *.: .
Mflv_0325|M.gilvum_PYR-GCK DMSAFAAQPMAAQAGAAFDVSNIIADIIRLYGEVLRFLFEPIFNFLR---
Mvan_0415|M.vanbaalenii_PYR-1 DVTAFAAQAQAAEAGSALDLSNIIADLIRAYGELLRFLFEPIFDFLR---
MSMEG_0619|M.smegmatis_MC2_155 SVQQMSAQATAAESGSSLDLSSIISDLLNQYLQGVPGG-QAILDFLK---
MAB_0809c|M.abscessus_ATCC_199 --HDHGHDDHDHEHGHDDDHG---------HGDLDPTDPEWWVHVAG---
TH_3113|M.thermoresistible__bu TGAHVTAELRAAESRTLLDEINELIDRLFAPLFPPGFADALPDGLLGTLL
MMAR_1028|M.marinum_M NGAPATTVTAAVTALRLPADASTVNFGLANIGSGNVGNANNGIGNLGSGN
MUL_0782|M.ulcerans_Agy99 NGAPATTVTAAVTALRLPADASTVNFGLANIGSGNVGNANNGIGNLGSGN
Mb1782c|M.bovis_AF2122/97 SGAPAAAMTAAAGIPALAGGPTAINLGIANVGGGNVGNANNGLANIGNAN
Rv1753c|M.tuberculosis_H37Rv SGAPAAAMTAAAGIPALAGGPTAINLGIANVGGGNVGNANNGLANIGNAN
MAV_4704|M.avium_104 FSNASTAVTRPAGSGAVARGYTSRIAGFLAPRAPQ---------------
MLBr_01182|M.leprae_Br4923 QEVVEEVVEEVVEEVVEAEQAISQAALDQAVNEGMEATVVPQVDQQVN--
Mflv_0325|M.gilvum_PYR-GCK -----------DPIGNTFALITDFLTNPVQALITWGPFLLAVVYQA----
Mvan_0415|M.vanbaalenii_PYR-1 -----------DPLGNTIKLITDFLTNPAQALITWGPFLAAVAYQA----
MSMEG_0619|M.smegmatis_MC2_155 -----------DPLGNLGRLLTDFMTNPAGALATWWPFLFGVAYQVFFNL
MAB_0809c|M.abscessus_ATCC_199 -----------EMVEHFELLLNNLLTDPAALLTNLPMVLADVAFHA----
TH_3113|M.thermoresistible__bu APLLPGATGDGPPMATIMSFLLMGLSNPAYLFPVLVYLLFTGVVHTTVAV
MMAR_1028|M.marinum_M LGNFNLGSGNLGSANAGWANLGSNNIGGANSGGNNLGWGNLGGLNTG-FA
MUL_0782|M.ulcerans_Agy99 LGNFNLGSGNLGSSNAGWANLGSNNIGGANSGGNNLGWGNLGGLNTG-FA
Mb1782c|M.bovis_AF2122/97 LGNYNFGSGNFGNSNIGSASLGNNNIGFGNLGSNNVGVGNLGNLNTG-FA
Rv1753c|M.tuberculosis_H37Rv LGNYNFGSGNFGNSNIGSASLGNNNIGFGNLGSNNVGVGNLGNLNTG-FA
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 ---VDVATPQTAVPDSSSAAAPQLWGGFAQHLSPINDTLSMINNHAG-MA
Mflv_0325|M.gilvum_PYR-GCK VSWVGASILYPSLFLLPLLATT---LAIVAGVG--QHLLSLLNTPPAEEA
Mvan_0415|M.vanbaalenii_PYR-1 VSWVGASILYPSLLLLPLVATT---LAIVLGVG--AYLLENLPAPAEDAP
MSMEG_0619|M.smegmatis_MC2_155 VGWPTWGMIFASPFLIPALAALGI-TGLVLLID--HVLNNPVDVPAEDEA
MAB_0809c|M.abscessus_ATCC_199 AQLASTIGQFAPALIQPALALAIANLGWAAGFAGLAGIQPSPEILAVEPG
TH_3113|M.thermoresistible__bu SGMLAQLVPLMPALLPVAVAALGGLAGVWAGLSGLAGLAGLTAVPAADGG
MMAR_1028|M.marinum_M NAGSGNFGFANTGNNNIGIGLTGDNQVGIGGLN--SGVGNFGLFNSGTGN
MUL_0782|M.ulcerans_Agy99 NAGSGNFRFANTGNNNIGIGLTGDNQVGIGGLN--SGVGNFGLFNSGTGN
Mb1782c|M.bovis_AF2122/97 NTGLGNFGFGNTGNNNIGIGLTGNNQIGIGGLN--SGTGNFGLFNSGSGN
Rv1753c|M.tuberculosis_H37Rv NTGLGNFGFGNTGNNNIGIGLTGNNQIGIGGLN--SGTGNFGLFNSGSGN
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 NAGLSLVNGMGSAMKSLAPTTTKAAESAFKAMG--SAVQSTGRGLLGSSS
Mflv_0325|M.gilvum_PYR-GCK AEEPAPAQSAPSRPDQQSFPATMSAPPPPSGAPIAANTVSAGSPPAPGAP
Mvan_0415|M.vanbaalenii_PYR-1 AEEPAASSPAPTRADQPSPAIAVSAPPPPSSAAATVGTVATGTAPAPGAP
MSMEG_0619|M.smegmatis_MC2_155 PQEVAAAP----RTDQP-LAVGISPAGVPGSAATAAAPAPAPAGTTVAAP
MAB_0809c|M.abscessus_ATCC_199 SVEDAPMAASAGTALAPSPPASAAAPAAPASAPAPTASGTAPAPPSP-PP
TH_3113|M.thermoresistible__bu ASAAPVPAGGPSLPSGSSAPPAPTGSAAPTTGTAPGGAGSPPPAGHSVPP
MMAR_1028|M.marinum_M VGLFNSGTGNFGIGNSGAYGTGLFNSGQANTGLLNAGSFNTGLLDVGNAN
MUL_0782|M.ulcerans_Agy99 VGLFNSGTGNFGIGNSGAYGTGLFNSGQANTGLLNAGSFNTGLLDVGNAN
Mb1782c|M.bovis_AF2122/97 VGFFNSGNGNFGIGNSGNFNTGGWNSGHGNTGFFNAGSFNTGMLDVGNAN
Rv1753c|M.tuberculosis_H37Rv VGFFNSGNGNFGIGNSGNFNTGGWNSGHGNTGFFNAGSFNTGMLDVGNAN
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 GGHVTAQLGRAASIGSLRVPQTWTTASQPVTAATRALSPARVAVATESES
Mflv_0325|M.gilvum_PYR-GCK AAAAATFVPYVVAGRD-PYEGFSPTVRDSTGAKAPAGTIPAAASGIAASA
Mvan_0415|M.vanbaalenii_PYR-1 AAATASFVPYAVAGRD-PGTGFSPTVRDSTSAKAPASGIPAAASGVAASA
MSMEG_0619|M.smegmatis_MC2_155 APAAATFVPYAVANRD-PGEGFTPTLGNKSQVAAPASGVPAAASGVAASL
MAB_0809c|M.abscessus_ATCC_199 VGDPGFSFPYAVGGGPRLGTGLALAAQVRRGTSAAARSSAREQAAAAETA
TH_3113|M.thermoresistible__bu APHSAPPVGTSGPPYLIAAQPAPGVARRSAGARRAASIVEPAGSAAAAAA
MMAR_1028|M.marinum_M TGSLNTGSFNMGSFNPGPSNTGALNTGTANTGWFNTGSLNTGAFNIGDMN
MUL_0782|M.ulcerans_Agy99 TGSLNTGSFNMGSFNPGPSNTGALNTGTANTGWFNTGSLNTGPFNIGDMN
Mb1782c|M.bovis_AF2122/97 TGSLNTGSYNMGDFNPGSSNTGTFNTGNANTGFLNAGNINTGVFNIGHMN
Rv1753c|M.tuberculosis_H37Rv TGSLNTGSYNMGDFNPGSSNTGTFNTGNANTGFLNAGNINTGVFNIGHMN
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 APLLGGGLPMAPMVPGGGSGTGGVNTALR---------------------
Mflv_0325|M.gilvum_PYR-GCK AERRKRRRRQKEEIKGRGYADAYADYEELPDDGPPEQPAPRVTASTRGAG
Mvan_0415|M.vanbaalenii_PYR-1 AERRKRRRRQKDEIAGRAYADAYADYEPEPDDEPPVRQEPRIAATERGAG
MSMEG_0619|M.smegmatis_MC2_155 ASRRKRRRKKGAQIEERGYADAYMDYEE-PDEDPAPQPEPRVRASERGSD
MAB_0809c|M.abscessus_ATCC_199 AARQARRRRDKGKQQGRG-----IEYMDMNATARCEEPAPDTTASHRGAG
TH_3113|M.thermoresistible__bu ARESAPIRRRRGQKDGLHG--------HRHEHQQPASMSAATAPSAAGIR
MMAR_1028|M.marinum_M NGLFNTGEMNNGIFYRGVGQGNFYFSITTPDLTLPPLEIPGITIPNLSLP
MUL_0782|M.ulcerans_Agy99 NGLFNTGEMNNGIFYRGVGQGNFYFSITTPDLTLPPLEIPGITIPNLSLP
Mb1782c|M.bovis_AF2122/97 NGLFNTGDMNNGVFYRGVGQGSLQFSITTPDLTLPPLQIPGISVPAFSLP
Rv1753c|M.tuberculosis_H37Rv NGLFNTGDMNNGVFYRGVGQGSLQFSITTPDLTLPPLQIPGISVPAFSLP
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------LQPRAFVMPRNPAAG---------
Mflv_0325|M.gilvum_PYR-GCK PMGFAGTVSRGDEQAGGLTTLA-GDSFGGGPREPMLPGTWDPDATDDRHT
Mvan_0415|M.vanbaalenii_PYR-1 PMGFAGTVSRDAAQAGGLTTLP-GDPFGGGPKAPMLPGTWDPDTEPD---
MSMEG_0619|M.smegmatis_MC2_155 AMGFTGTVTRHDVTAAGLTTLT-GDTFDDRPVTPMLPGTWDGDENPAP--
MAB_0809c|M.abscessus_ATCC_199 TMGFAGTVIKTGATASGLAMLSRDDDLFEGVRAPMVPNTWDP--------
TH_3113|M.thermoresistible__bu VLGGAGRISRTRGSVPHAEITP-GDPTEPSSPTDRAHRPAEPN-------
MMAR_1028|M.marinum_M PITVPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGGFD
MUL_0782|M.ulcerans_Agy99 PITMRR--------------------------------------------
Mb1782c|M.bovis_AF2122/97 AITLPSLTIPAATTPANITVGAFSLPGLTLPSLNIPAATTPANITVGAFS
Rv1753c|M.tuberculosis_H37Rv AITLPSLNIPAATTPANITVGAFSLPGLTLPSLNIPAATTPANITVGAFS
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK DGRDGHDGHDGKEKP-----------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 ---ERHNHHDGKDSQ-----------------------------------
MSMEG_0619|M.smegmatis_MC2_155 ----SPDDPEGGPRR-----------------------------------
MAB_0809c|M.abscessus_ATCC_199 ----EHGRQD----------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M LPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVVGG
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 LPGLTLPSLNIPAATTPANITVGAFSLP----------------------
Rv1753c|M.tuberculosis_H37Rv LPGLTLPSLNIPAATTPANITVGAFSLPGLTLPSLNIPAATTPANITVGA
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M FDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANVVV
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 ------------------------------GLTLPSLNIPAATTPANITV
Rv1753c|M.tuberculosis_H37Rv FSLPGLTLPSLNIPAATTPANITVGAFSLPGLTLPSLNIPAATTPANITV
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M GGFDLPGVTLPALTIPAATTPANVVVGGFDLPGVTLPALTIPAATTPANV
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 SGFQLPPLSIPSVAIPPVTVPP----------------------------
Rv1753c|M.tuberculosis_H37Rv SGFQLPPLSIPSVAIPPVTVPP----------------------------
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M VVGGFDLPGVTLPALTIPAATTPANVVVGGFDLPQLGIPSLTIPPLTLAE
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 --------------------------------------------------
Rv1753c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M GTTVGAFSLPELGIPSLTIPPLTLAEGTTVGAFSLPELGIPSLTIPPLTL
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 --------------------------------------------------
Rv1753c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M AEGTTVGAFSLPQLGIPSLTIPSLTVPAGTTVGGFDLPALQTRSITVGGI
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 ---ITVGAFNLPPLQIPEVTIPQLTIPAGITIGGFSLPAIHTQPITVGQI
Rv1753c|M.tuberculosis_H37Rv ---ITVGAFNLPPLQIPEVTIPQLTIPAGITIGGFSLPAIHTQPITVGQI
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M DAGGFRTPAISTVSSTGYLELPTVTIPGVRVNNFVAFIPNNIEFLQTGQP
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 GVGQFGLPSIG---------------------------------------
Rv1753c|M.tuberculosis_H37Rv GVGQFGLPSIG---------------------------------------
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M GVIPQIGGLEHQAYISTGDIVIQGFSISGIGFSLPQIDVGAFTLPELTIP
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 ----------WDVFLST---------------------------PRITVP
Rv1753c|M.tuberculosis_H37Rv ----------WDVFLST---------------------------PRITVP
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M PIGIPPTIVGAFTLPEITTPVITTPPLVVDPVSVAGFTLPEITTPEFTTP
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 AFGIP------FTLQFQTN----VPALQPPGGGLSTFTNGALIFGEFDLP
Rv1753c|M.tuberculosis_H37Rv AFGIP------FTLQFQTN----VPALQPPGGGLSTFTNGALIFGEFDLP
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M PLVVDPVSVAGFTLPEITTPEFTTPPLVVDPVSVAGFTLPEITTPEFTTP
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 QLVVHPYTLTG--------------PIVIGSFFLPAFNIPGIDVP-----
Rv1753c|M.tuberculosis_H37Rv QLVVHPYTLTG--------------PIVIGSFFLPAFNIPGIDVP-----
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M PLVVDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTA
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 --------------------------------------------------
Rv1753c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M PFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTA
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 -----AINVDGFTLPQITTPAITTPEFAIPPIGVGGFTLPQITTQEIITP
Rv1753c|M.tuberculosis_H37Rv -----AINVDGFTLPQITTPAITTPEFAIPPIGVGGFTLPQITTQEIITP
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M PFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTA
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 ELTINSIGVGGFTLPQITTPPITTPPLTIDPINLTGFTLPQITTPPITTP
Rv1753c|M.tuberculosis_H37Rv ELTINSIGVG----------------------------------------
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M PFTIDPVSVAGFTLPQISTPEFTTAPFTIDPVSVAGFTLPQISTPEFTTA
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 PLTIDPINLTGFTLPQITTPPITTPPLTIDPINLTGFTLPQITTPPITTP
Rv1753c|M.tuberculosis_H37Rv ----------GFTLPQITTPPITTPPLTIDPINLTGFTLPQITTPPITTP
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M PFTIDPVSVAGFTLPQISTPQFTTAPITIPPIGLGSFTTPPLTIPSIHLP
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 PLTIDPINLTGFTLPQITTPPITTPPLTIEPIGVGGFTTPPLTVPGIHLP
Rv1753c|M.tuberculosis_H37Rv PLTIDPINLTGFTLPQITTPPITTPPLTIEPIGVGGFTTPPLTVPGIHLP
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M GTTIGEFAFPAGPGYLNSSATPSSGFFNSGAGGNSGLGNNGSGLSGWFNT
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 STTIGAFAIPGGPGYFNSSTAPSSGFFNSGAGGNSGFGNNGSGLSGWFNT
Rv1753c|M.tuberculosis_H37Rv STTIGAFAIPGGPGYFNSSTAPSSGFFNSGAGGNSGFGNNGSGLSGWFNT
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0619|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0809c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3113|M.thermoresistible__bu --------------------------------------------------
MMAR_1028|M.marinum_M NPVGLLGGSGYQNYGGLSSGFSNLGSGVSGFANTGVRPFSLTGLISGFAN
MUL_0782|M.ulcerans_Agy99 --------------------------------------------------
Mb1782c|M.bovis_AF2122/97 NPAGLLGGSGYQNFGGLSSGFSNLGSGVSGFANRGILPFSVASVVSGFAN
Rv1753c|M.tuberculosis_H37Rv NPAGLLGGSGYQNFGGLSSGFSNLGSGVSGFANRGILPFSVASVVSGFAN
MAV_4704|M.avium_104 --------------------------------------------------
MLBr_01182|M.leprae_Br4923 --------------------------------------------------
Mflv_0325|M.gilvum_PYR-GCK --------------
Mvan_0415|M.vanbaalenii_PYR-1 --------------
MSMEG_0619|M.smegmatis_MC2_155 --------------
MAB_0809c|M.abscessus_ATCC_199 --------------
TH_3113|M.thermoresistible__bu --------------
MMAR_1028|M.marinum_M IGSNLSGFFGSNVP
MUL_0782|M.ulcerans_Agy99 --------------
Mb1782c|M.bovis_AF2122/97 IGTNLAGFFQGTTS
Rv1753c|M.tuberculosis_H37Rv IGTNLAGFFQGTTS
MAV_4704|M.avium_104 --------------
MLBr_01182|M.leprae_Br4923 --------------