For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NTDDMILISVDDHIVEPPDMFDNHLPAKYLRDAPRLVRNPDGSDVWKFRDSVIPNPALNAVAGRPKEEYG LEPQGLDEIRPGCYQVDERVKDMNAGGILASICFPSFPGFAGRRFATDDPDFSLALIQAYNDWHIDQWCG AYPARFIPMALPVIWDAQACATEVRRVSKKGVHALTFTENPATMGYPSFHNDYWNPLWKALCDTNTVMNI HFGSSGNLVTTAPDAPIDVLMTLGPMNIVQAAADLLWSRPIKDYPDLKIGLSEGGTGWIPYFLERADRVF EMHSTWTHQDFGGKLPSEVFREHFLACFISDPVGVKLRNMIGIDNIAWEADYPHSDSMWPGAPEELGEVL TANSVPDLEVDKMTHLNAMRWYSFDPFSRIPREQATVGALRKAAAGHDVSTRPRSQHKSGERTSILVEP*
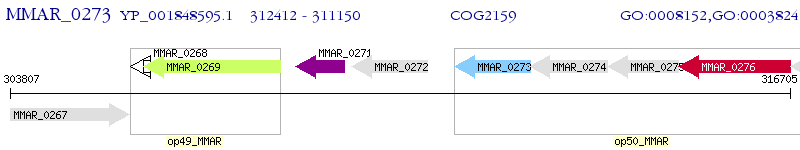
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0273 | - | - | 100% (420) | amidohydrolase |
| M. marinum M | MMAR_4742 | - | 0.0 | 84.25% (419) | metal-dependent amidohydrolase |
| M. marinum M | MMAR_2618 | - | 7e-87 | 44.24% (373) | amidohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4324 | - | 0.0 | 78.26% (414) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_0907 | - | 0.0 | 81.86% (419) | amidohydrolase 2 |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 0.0 | 76.47% (408) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2339 | - | 1e-118 | 47.79% (408) | amidohydrolase 2 |
| M. ulcerans Agy99 | MUL_3143 | - | 3e-85 | 43.97% (373) | amidohydrolase |
| M. vanbaalenii PYR-1 | Mvan_2022 | - | 0.0 | 78.50% (414) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0273|M.marinum_M -----MNTDDMILISVDDHIVEPPDMFDNHLPAKYLRDAPRLVRNPDGSD
MAV_0907|M.avium_104 -----MNKEDMILISVDDHTVEPPDMFKNHLPKKYLDDAPRLVHNPDGSD
Mflv_4324|M.gilvum_PYR-GCK -----MNRDDMILISVDDHIVEPPDMFKNHLSKKYLVEAPRLVHNEDGSD
Mvan_2022|M.vanbaalenii_PYR-1 -----MNRDDMILISVDDHIVEPPDMFKNHLPSKYLDEAPRLVHNPDGSD
MSMEG_4837|M.smegmatis_MC2_155 -----MNKDDMILISVDDHIIEPPDMFKNHLPAKYRDEAPRLVHNPDGSD
TH_2339|M.thermoresistible__bu -MSHDLRPEDLILVSIDDHVVEPPDMFLRHVPAKYRDEAPIVVTDENGVD
MUL_3143|M.ulcerans_Agy99 MTDPPRARRRYTVISVDDHIVEPPDTFTGRLPQRFADRAPRVVETDDGGQ
::*:*** :**** * ::. :: ** :* :* :
MMAR_0273|M.marinum_M VWKFRDSVIPNPALNAVAGRPKEEYGLEPQGLDEIRPGCYQVDERVKDMN
MAV_0907|M.avium_104 MWKFRDTVIPNVALNAVAGRPKEEYGIEPTGLDEIRPGCYNVDERVKDMN
Mflv_4324|M.gilvum_PYR-GCK TWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGCWQVDERVKDMN
Mvan_2022|M.vanbaalenii_PYR-1 TWQFRDVVIPNVALNAVAGRPKEEYGLEPQGLDEIRPGCWQVDERVKDMN
MSMEG_4837|M.smegmatis_MC2_155 TWQFRDTVIPNVALNAVAGRPKEEYGLEPQGLDEIRKGCYDPNERVKDMN
TH_2339|M.thermoresistible__bu QWMYQGRPQGVSGLNAVVSWPPEEWGRNPARFAEMRPGVYDVHERVRDMN
MUL_3143|M.ulcerans_Agy99 VWVYDGQVLPNVGFNAVVGRPVSEYGFEPVRFDEMRRGAWDIHERIKDMD
* : . .:***.. * .*:* :* : *:* * :: .**::**:
MMAR_0273|M.marinum_M AGGILASICFPSFP-GFAGR--RFATDDPDFSLALIQAYNDWHIDQWCGA
MAV_0907|M.avium_104 AGGILASICFPSFP-GFAGR--LFATEDNDFSIALVQAYNDWHIDEWCGA
Mflv_4324|M.gilvum_PYR-GCK AGGILGSMCFPSFP-GFAGR--LFATEDPEFSLALVQAYNDWHVEEWCGA
Mvan_2022|M.vanbaalenii_PYR-1 AGGILGSMCFPSFP-GFAGR--LFATEDAEFSLALVQAYNDWHVEEWCGA
MSMEG_4837|M.smegmatis_MC2_155 AGGVLATMNFPSFP-GFAAR--LFATDDEEFSLALVQAYNDWHIDEWCAS
TH_2339|M.thermoresistible__bu RNGILASMCFPTFT-GFSAR--HLNMHREHVTLIMVSAYNDWHIDEWAGS
MUL_3143|M.ulcerans_Agy99 LNGVYASLNFPSFLPGFAGQRLQQVTKDPELAWAAVRAWNDWHLEAWAGP
.*: .:: **:* **:.: . ..: : *:****:: *...
MMAR_0273|M.marinum_M YPARFIPMALPVIWDAQACATEVRRVSKKGVHALTFTENPATMGYPSFHN
MAV_0907|M.avium_104 YPARFIPMAIPVIWDAEECAKEVRRVSKKGVHALTFTENPAAMGYPSFHD
Mflv_4324|M.gilvum_PYR-GCK YPARFIPMTLPVIWDPEACAKEIRRNAARGVHSLTFSENPSAMGYPSFHD
Mvan_2022|M.vanbaalenii_PYR-1 YPARFIPMTLPVIWDPVACAAEIRRNAARGVHSLTFSENPSAMGYPSFHD
MSMEG_4837|M.smegmatis_MC2_155 NPGRFIPMAIPAIWNPGLAAAEVRRVAEKGVHSLTFTENPSTLGYPSFHD
TH_2339|M.thermoresistible__bu YPGRFIPIAILPTWNPEAMCAEIRRVAAKGCRAVTMPELPHLEGLPSYHD
MUL_3143|M.ulcerans_Agy99 YPERIIPCQLPWLLDPSAGAQLIHENAERGFRAVSFSENPAMLGLPSIHS
* *:** : :. . ::. : :* :::::.* * * ** *.
MMAR_0273|M.marinum_M -DYWNPLWKALCDTNTVMNIHFGSSGNLVTTAPDAPIDVLMTLGPMNIVQ
MAV_0907|M.avium_104 -EYWNPLWKALVDTNTVMNVHIGSSGRLAITAPDAPMDVMITLQPMNIVQ
Mflv_4324|M.gilvum_PYR-GCK FDHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDVMITLQPMNIVQ
Mvan_2022|M.vanbaalenii_PYR-1 FEHWKPMWDALVDTDTVLNVHIGSSGRLAITAPDAPMDVMITLQPMNIVQ
MSMEG_4837|M.smegmatis_MC2_155 LEYWRPLWQALVDTETVMNVHIGSSGKLAITAPDAPMDVMITLQPMNIVQ
TH_2339|M.thermoresistible__bu EDYWGPVFRTLSEENVVMCLHIGTGFGAISMAPDAPIDNMIILATQVSAM
MUL_3143|M.ulcerans_Agy99 -GHWDPMMAACAETGTVVNLHIGSSGSSPSTTDDAPADVPGVLFFAYAIS
:* *: : : .*: :*:*:. : *** * *
MMAR_0273|M.marinum_M AAADLLWSRPIKDYPDLKIGLSEGGTGWIPYFLERADRVFEMHSTWTH-Q
MAV_0907|M.avium_104 AAADLLWSRPIKEFPDLKIALSEGGTGWIPYFLERADRTFEMHSAWTH-Q
Mflv_4324|M.gilvum_PYR-GCK AAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERVDRTYEMHSTWTG-Q
Mvan_2022|M.vanbaalenii_PYR-1 AAADLLWSRPIKEYPDLKIALSEGGTGWIPYFLERVDRTYEMHSTWTG-Q
MSMEG_4837|M.smegmatis_MC2_155 AAADLLWSAPIKEFPTLKIALSEGGTGWIPYFLDRVDRTFEMHSTWTH-Q
TH_2339|M.thermoresistible__bu CAQDLLWGPAMRNYPDLKFAFSEGGIGWIPFYLDRCDRHYTN-QKWLR-R
MUL_3143|M.ulcerans_Agy99 AAVDWLYSGLPSRFPDLKICLSEGRIGWVAGLLDRLDHMLSYHEMYGTWR
.* * *:. :* **: :*** **:. *:* *: . : :
MMAR_0273|M.marinum_M DFGGKLP-SEVFREHFLACFISDPVGVKLRNMIGIDNIAWEADYPHSDSM
MAV_0907|M.avium_104 DFKGKLP-SEVFREHFLTCFISDKVGVALRNEIGIDNICWEADYPHSDSM
Mflv_4324|M.gilvum_PYR-GCK DFKGKLP-SEVFREHFLTCFIADPVGVATRHQIGVDNICWEADYPHSDSM
Mvan_2022|M.vanbaalenii_PYR-1 DFKGKLP-SEVFREHFLTCFIADPVGVATRHQIGVDNICWEADYPHSDSM
MSMEG_4837|M.smegmatis_MC2_155 NFGGKLP-SEVFREHFMTCFISDPVGVKNRHMIGIDNICWEMDYPHSDSM
TH_2339|M.thermoresistible__bu DFGDKLP-SDVFREHSLACYVTDKTSLKLRHEIGIDIIAWECDYPHSDCF
MUL_3143|M.ulcerans_Agy99 AMGESLTSAEVFTRNFWFCAVEDKSSFVQHYRIGTENIMVESDYPHCDST
: .*. ::** .: * : * .. : ** : * * ****.*.
MMAR_0273|M.marinum_M WPGAPEELGEVLTANSVPDLEVDKMTHLNAMRWYSFDPFSRIPREQATVG
MAV_0907|M.avium_104 WPGAPEELWDVLSINNVPDDEINKMTYQNAMRWYSFDPFTHISREQATVG
Mflv_4324|M.gilvum_PYR-GCK WPGAPEQLDEVLKANNVPDDEINKMTYENAMRWYHWDPFTHIAKEQATVG
Mvan_2022|M.vanbaalenii_PYR-1 WPGAPEQLDEVLKVNNVPDDEIDKMTYQNAMRWYHWDPFTHIPKEQATVG
MSMEG_4837|M.smegmatis_MC2_155 WPGAPEELSAVFDTYDVPDDEINKITHENAMRLYHFDPFAHVPKEQATVG
TH_2339|M.thermoresistible__bu WPDAPEQVHAELRAAGADDFDINKITWENSCRFFSWDPFEHTPRAEATVG
MUL_3143|M.ulcerans_Agy99 WPHTQQTMHEQLDG--LPADVIRKITWENAARLYQHPVPVAIAQNPEAF-
** : : : : : *:* *: * : .: :.
MMAR_0273|M.marinum_M ALRKAAAGHDVSTRPRSQHKSGERTSILVEP--------------
MAV_0907|M.avium_104 ALRKAAEGHDVSIKAQGHDKDSRGGSSFAEFAANAKALTGNKD--
Mflv_4324|M.gilvum_PYR-GCK ALRKAAEGHDVSIQALSHHKEGERGGGAGHFDALNAAARGNSGA-
Mvan_2022|M.vanbaalenii_PYR-1 ALRKAAEGHDVSIQALSHHKEGERGGGAGHFDALNAAARGNSGAD
MSMEG_4837|M.smegmatis_MC2_155 ALRKAAEGHDVSIRALS--KKDKTGASFADFQANAKAVSGARD--
TH_2339|M.thermoresistible__bu ALRAKATDVDTTIRSRA-----EWAQLYEQKQQFAQA--------
MUL_3143|M.ulcerans_Agy99 ---------------------------------------------