For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRTQLGRPVGASGEETRQRIIVATMQCVSKVGYARATIREIARTANVTSASLYNYFPNKSELIKETIAA RADAAMPRLRRAAEGGGNIVDRIEAVLDECGELIREYPQLAAFEFAIRAEDGIALDAQEGSGQLGEVGFT AFREIIRGLVEDARRRGELAEQADTAGAIEAIYALIYGLTELAATLPLDQFHAALSAAKGLIRGTLFRC
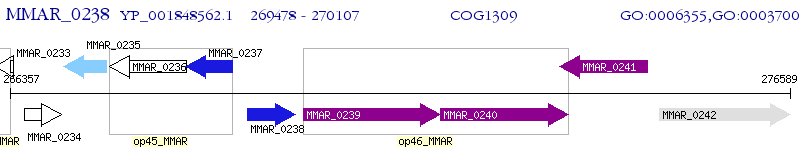
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0238 | - | - | 100% (209) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0237 | - | 1e-44 | 48.99% (198) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0355 | - | 4e-10 | 25.43% (173) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0149 | - | 9e-15 | 28.78% (205) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3637 | - | 7e-12 | 26.09% (184) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0144 | - | 9e-15 | 28.78% (205) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0579c | - | 1e-08 | 30.39% (102) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5151 | - | 2e-11 | 27.05% (207) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1565 | - | 1e-14 | 26.60% (188) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_3831 | - | 2e-11 | 28.98% (176) | PUTATIVE putative transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_4875 | - | 1e-113 | 99.52% (209) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2776 | - | 7e-14 | 28.90% (173) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0238|M.marinum_M --------------------------------------------------
MUL_4875|M.ulcerans_Agy99 --------------------------------------------------
Mb0149|M.bovis_AF2122/97 MPHSWTPTSVMTPPLVVAAFRPVGHYRLATDRAGGPCSPPATGAKLTSSV
Rv0144|M.tuberculosis_H37Rv MPHSWTPTSVMTPPLVVAAFRPVGHYRLATDRAGGPCSPPATGAKLTSSV
MAV_5151|M.avium_104 --------------------------------------------------
TH_3831|M.thermoresistible__bu --------------------------------------------------
Mflv_3637|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2776|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1565|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0579c|M.abscessus_ATCC_199 ---------------------------------------------MATPR
MMAR_0238|M.marinum_M ----------------------MTRTQLGRPVGASGE-ETRQRIIVATMQ
MUL_4875|M.ulcerans_Agy99 ----------------------MTRTQLGRPVGASGE-ETRQRIIVATMQ
Mb0149|M.bovis_AF2122/97 ASRPTVGTKPQWWHTLVMSMSLTAGRGPGRPPAAKAD-ETRKRILHAARQ
Rv0144|M.tuberculosis_H37Rv ASRPTVGTKPQWWHTLVMSMSLTAGRGPGRPPAAKAD-ETRKRILHAARQ
MAV_5151|M.avium_104 -------------------MLPRGNRRPGRPPAAKAD-ETRQRIIQAARL
TH_3831|M.thermoresistible__bu ----------------------------MRPARTASS-DTRARILEAARR
Mflv_3637|M.gilvum_PYR-GCK --------------MENETLSVSAGRKRGRPVAADSE-QTRRTVLAAARD
Mvan_2776|M.vanbaalenii_PYR-1 ---------------------MPAVRRPGRPVGANSD-QTRRTILCAARD
MSMEG_1565|M.smegmatis_MC2_155 -----------------MRQTRAARSGPGRPAGTDSV-DTRQRVIDAACR
MAB_0579c|M.abscessus_ATCC_199 TSSASEAAQSGDGDTDATSQPAAVSALSDDDLGSSAQRERRKRILDATLA
: . : * :: *:
MMAR_0238|M.marinum_M CVSKVGYARATIREIARTANVTSASLYNYFPNKSELIKETIAARADAAMP
MUL_4875|M.ulcerans_Agy99 CVSKVGYARATIREIARTANVTSASLYNYFPNKSELIKETIAARADAAMP
Mb0149|M.bovis_AF2122/97 VFSERGYDGATFQEIAVRADLTRPAINHYFANKRVLYQEVVEQTHELVIV
Rv0144|M.tuberculosis_H37Rv VFSERGYDGATFQEIAVRADLTRPAINHYFANKRVLYQEVVEQTHELVIV
MAV_5151|M.avium_104 VFSERGYDGATFQAIAARADLTRPAINHYFSSKRALYQEVMDETNEFVIG
TH_3831|M.thermoresistible__bu IIVERGYPAATCQGIARAAGVSRPTLHYYFASREEIYDCLVEQSRNLLAE
Mflv_3637|M.gilvum_PYR-GCK IIAERGYQATTFQQIAQRAGVSRPTLHYYFATREDLYDALLADVCERLGR
Mvan_2776|M.vanbaalenii_PYR-1 VIAEHGYRAATFQQIALRAGVSRPTLHYYFATREQLYEVLLADVRAEVAR
MSMEG_1565|M.smegmatis_MC2_155 CFAQFGYGPATNNQIAEMAGVTAGSVYYHFGTKNKLFEAVCDDVYGKILT
MAB_0579c|M.abscessus_ATCC_199 IASKGGYEAVQMRTVAERADVAVGTLYRYFPSKVHLLVSALEREFEQIDA
: ** . . :* *.:: :: :* .: :
MMAR_0238|M.marinum_M RLRRAAEGGGNIVDRIEAVLDECGELIREYPQLAAFEFAIRAEDGIALDA
MUL_4875|M.ulcerans_Agy99 RLRRAAEGGGNIVDRIEAVLDECGELIREYPQLAAFEFAIRAEDGIAVDA
Mb0149|M.bovis_AF2122/97 AGIERARREPTLMGRLAVVVDFAMEADAQYPASTAFLATTVLESQ----R
Rv0144|M.tuberculosis_H37Rv AGIERARREPTLMGRLAVVVDFAMEADAQYPASTAFLATTVLESQ----R
MAV_5151|M.avium_104 VGIKEADRETTLVGRLTAFISAAVKANAENPAGSAFIIGGVLESQ----R
TH_3831|M.thermoresistible__bu C-AERAMRYDTLVERLTEFGLRLREVDSRDRSTIAFLISARLEGM----R
Mflv_3637|M.gilvum_PYR-GCK CAAAAAGQ-DSLRGQLAAFMAEMNRLAVAEPAMMKLVVTARIDHHGARIG
Mvan_2776|M.vanbaalenii_PYR-1 CAAEATLA-GSLLRQLEVFIAEMHRLGGAEPALMGLVVTARVDHRGVLR-
MSMEG_1565|M.smegmatis_MC2_155 RVMLAVSGSHSVVGLLRAVLTESMRINHESPELAGFVATAPIDAR----R
MAB_0579c|M.abscessus_ATCC_199 KSDRNAVSGGTPYERLHAAITRLNRNMQRNPLLTEAMTRALVFAD-----
. . : .
MMAR_0238|M.marinum_M QEGSGQLGEVGFTAFREIIRGLVEDARRRGELAEQADTAGAIEAIYALIY
MUL_4875|M.ulcerans_Agy99 QEGSGQLGEVGFTAFREIIRGLVEDARRRGELAEQADTAGAIEAIYALIY
Mb0149|M.bovis_AF2122/97 HPELSRTENDAVRATREFLVWAVNDAIERGELAADVDVSSLAETLLVVLC
Rv0144|M.tuberculosis_H37Rv HPELSRTENDAVRATREFLVWAVNDAIERGELAADVDVSSLAETLLVVLC
MAV_5151|M.avium_104 HPEWNTAENDSVRIAREFLIRVVNDAIEHGEVAADIDASALVETLLVVMC
TH_3831|M.thermoresistible__bu NRELG---EDPAMALREFFTELLADAVAAGELPSDIDIESAVEVMYTVLW
Mflv_3637|M.gilvum_PYR-GCK TGEGRGAATDIVSTVHDFYDAVVLAAAARGELPAGHDPHAVADMLAAVFW
Mvan_2776|M.vanbaalenii_PYR-1 ----HEAATAVVATVHAFFDAVVVDAVRRGELAFDTDAHAVADLLGALFW
MSMEG_1565|M.smegmatis_MC2_155 HRELAESFATQGARMADALTDAVRSGQDAGDIAADLDPVRVARLISAVVD
MAB_0579c|M.abscessus_ATCC_199 -----ASAAGEVDHVGRLMDGIFARAMAGEDDPTDAQFHIARVISDVWTS
. . : . : .
MMAR_0238|M.marinum_M GLTELAATLP-LDQFHAALSAAKGLIRGTLFRC-----------
MUL_4875|M.ulcerans_Agy99 GLTELAATLP-LDQFHAALSAAKGLIRGTLFRC-----------
Mb0149|M.bovis_AF2122/97 GVGFYIGFVGSYQRMATITDSFQQLLAGTLWRPPT---------
Rv0144|M.tuberculosis_H37Rv GVGFYIGFVGSYQRMATITDSFQQLLAGTLWRPPT---------
MAV_5151|M.avium_104 GVGLYAGYVETYQEMLAVTGMLRQLLEGALWRPGS---------
TH_3831|M.thermoresistible__bu GLGFYTGFVERSTDMLVLAEQLENLFASA---------------
Mflv_3637|M.gilvum_PYR-GCK GLGFHARMLTADIGVDGVPEVARQLLSVLDSGLLDGALRAPADA
Mvan_2776|M.vanbaalenii_PYR-1 GLVFHAGFIS---GADGAPEIARQLLRMVGFGLLDTPEPASVDA
MSMEG_1565|M.smegmatis_MC2_155 GFAHAAVSAD-PDEMDNMNELFQSLLLDTT--------------
MAB_0579c|M.abscessus_ATCC_199 NMIAWLTRRASATDVSHRLDRTVRLLLGIT--------------
.. *: