For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LGKGYWFDEVYMLAIGRFHLDWGAADQPPVTPALAALADTVAPGSLVTLRAPAVLATGCAVVLAALIARE LGGRRRAQAVTALAQATALFSTFAGHWLTPYTLEPAQWLLLIWLLVRWIRTRDDRLLVAAGAATGLAAMT KFQVLLLCAVLLLGIAWCGPRALLRRPLLWVGAAVALAMVSPTLVWQHVHGWPQLQMTTVVAGEAEYLYG GRAGVAVQLILFAGVLGVGLGVYGAWCLLRHAEFREYRFLPAVGVVLYVVFVATAGRPYYLGGLYAPFVA AGAVCLESVSPGTVRRWALRVGTVTAVAATVAILVLSVNITPPDVGDRIAKAAATAYRDLPEDERDRTVL LGESYITAAYLDGAAPRYGLPQAYGTNRSYGYFTPPPDTADEALYVGAEPDGMREHFVTVRNLATIDGDL KLFLLRERRHPWPQIWSEQRSLTVS
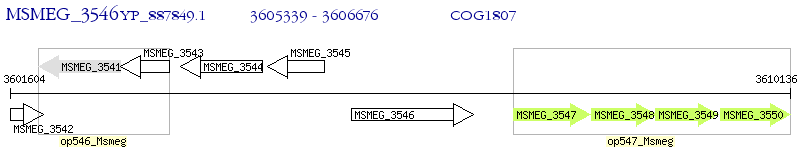
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3546 | - | - | 100% (445) | hypothetical protein MSMEG_3546 |
| M. smegmatis MC2 155 | MSMEG_2315 | - | 3e-81 | 40.47% (467) | hypothetical protein MSMEG_2315 |
| M. smegmatis MC2 155 | MSMEG_1035 | - | 3e-81 | 40.47% (467) | hypothetical protein MSMEG_1035 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2269 | - | 6e-05 | 23.76% (202) | glycosyl transferase family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01389 | - | 2e-05 | 23.38% (154) | hypothetical protein MLBr_01389 |
| M. abscessus ATCC 19977 | MAB_1277 | - | 3e-78 | 37.95% (477) | hypothetical protein MAB_1277 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3138 | - | 4e-05 | 24.22% (289) | hypothetical protein MAV_3138 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3546|M.smegmatis_MC2_155 ------------------------------------------------MG
MAB_1277|M.abscessus_ATCC_1997 ------MAAVTSRYDDGNNIPGVPPVCIARADLTAITTLTFLVFGWASRG
MLBr_01389|M.leprae_Br4923 MTSIMSVSALEQSAADVGDNSARQHAHGALPDSLAIAMLATVISGAWASR
MAV_3138|M.avium_104 -----------------------------MLDPWAIAVLATALSAAWACR
Mflv_2269|M.gilvum_PYR-GCK ------MTTTVRTGVRSAPHPEADTRPRWVTPALLALLASTAVLYLWGLG
MSMEG_3546|M.smegmatis_MC2_155 KGYWFDEVYMLAIGRFHLD---WGAADQPP------VTPALAALADT---
MAB_1277|M.abscessus_ATCC_1997 YGYFFDEAYFVVAGRDHLS---WGYFDQPP------LVPFIAGVADN---
MLBr_01389|M.leprae_Br4923 PSLWFDEAATISASASRTVPELWRLLSHID------AVHGLYYLLMHGWF
MAV_3138|M.avium_104 PSLWFDEGATISAAASRTLPELWRLLGHID------AVHGAYYLLMHGWF
Mflv_2269|M.gilvum_PYR-GCK SSGWANQYYAAAAQAGTQDWKAWLFGSLDAGNAITVDKPPASLWVMALSG
. : :: * .
MSMEG_3546|M.smegmatis_MC2_155 -VAPGSLVTLRAPAVLATGCAVVLAALIARELGGRRRAQAVTALAQATAL
MAB_1277|M.abscessus_ATCC_1997 -IAPGSLWVLRLAATAAVAIGTLMCGLIAAELGAGRLTQSLSAIAHATAM
MLBr_01389|M.leprae_Br4923 AIFPSTEFWSRVPSCLAIGAAAAGVTVFTRQFAT-RTTAVYAGIVFAILP
MAV_3138|M.avium_104 ALFPPTEFFSRLPSALAVGAAAAGVTVFTRQFAP-RRTAVCAGAVFALLP
Mflv_2269|M.gilvum_PYR-GCK RVFGFSPFTMLLPQALMGVASVAVLVAAVRRLSG-PGAGLIAGAALAVTP
: : . .. . .:. : :. . *
MSMEG_3546|M.smegmatis_MC2_155 FSTFAGHWLTPYTLEPAQWLLLIWLLVRWIRTR-----DDRLLVAAGAAT
MAB_1277|M.abscessus_ATCC_1997 FQILA-HWLQTPAIDPALWAVVCWLLVRWTRSLRERLPDDRLLLWAGIVT
MLBr_01389|M.leprae_Br4923 RITWAGIEARSSALSVAAAMWLTVLLVASVQRN-----RPRLWLCYALTL
MAV_3138|M.avium_104 RMTWAGMEARPYAFVAAAAVWLTVLFVAAVRRG-----APRGWVGYALAL
Mflv_2269|M.gilvum_PYR-GCK VAALMFRYNNPDALLVLLLVVAAYLMVRAIQTG-----GTRWVVLVGVVL
. :: *:* : * : . .
MSMEG_3546|M.smegmatis_MC2_155 GLAAMTKFQVLLLCAV-LLLGIAWCGPRALLRRP--------LLWVG--A
MAB_1277|M.abscessus_ATCC_1997 AISMQTKFLIPALWFA-VLLSAAVLGPRALLRRR--------QLWIG--A
MLBr_01389|M.leprae_Br4923 MLSILLNLTLATLVLV-YAVILPWLAPNKFRNSP--------FIWWAVTS
MAV_3138|M.avium_104 MLAILLNLNMVLMVPV-YGVMLPLLTARGARRSA--------ALWWAGSS
Mflv_2269|M.gilvum_PYR-GCK GVAFLTKMLQAFLVVPGLALAFLVAAPVGVWARVGKLAAGGVAMIATAGS
:: :: : : . : :
MSMEG_3546|M.smegmatis_MC2_155 AVALAMVSPTLVWQHVHGWPQLQMTTVVAG--------------------
MAB_1277|M.abscessus_ATCC_1997 AVVVVATAPTLIWQAAHGWPYLRMAAIVAA--------------------
MLBr_01389|M.leprae_Br4923 VVALGTITPFILFAHGQVWQVDWIFRVSWHY-------------------
MAV_3138|M.avium_104 AVAVGAMTPFLLFAHNQVWQVNWIYPVSWHY-------------------
Mflv_2269|M.gilvum_PYR-GCK FLTLVSVWPADARPYIGGSTDNSLLQLALGYNGIQRVMGGQGGPGGPPGG
:.: * : :
MSMEG_3546|M.smegmatis_MC2_155 -------------------------EAEYLYGGRAGVAVQLILFAGVLGV
MAB_1277|M.abscessus_ATCC_1997 -------------------------ESE----RGAALLGGMLVGSAVIGL
MLBr_01389|M.leprae_Br4923 -------------------------VFDITQRQYFDHSVSFAIATAVIIV
MAV_3138|M.avium_104 -------------------------AFDIILRQYFDHSVALAVLSAVLIV
Mflv_2269|M.gilvum_PYR-GCK GLGGPPAGGPGGPGGGANLFFGGEPDIGRLFGASMGAEASWLLPAALIGL
: :.:: :
MSMEG_3546|M.smegmatis_MC2_155 GLGVYGAWCLLRH-AEFREYRFLPAVGVVLYVVFVATAG---RPYYLGGL
MAB_1277|M.abscessus_ATCC_1997 VLLLAGIVGLLAS-ASLRPYRYLGSAAVLVGAFMFASHG---RAHYAMGL
MLBr_01389|M.leprae_Br4923 PAIATRLAGLRAPAGDLRSLVIICTAWIVIPTTLMVGYSAVIEPVYYPRY
MAV_3138|M.avium_104 AAAVARLAGVPAPPGDLRRLLILCAAWMVIPTALVVVYSAVAEPIYYPRY
Mflv_2269|M.gilvum_PYR-GCK VAALWITRRAART-DAIRAGLLLWGGWVLVTGVVFSYMDGIIHPYYTVAL
:* : ::: .. . .. *
MSMEG_3546|M.smegmatis_MC2_155 YAPFVAAGAVCLESVSPG-TVRRWALR--VGTVTAVAATVAILVLSVN--
MAB_1277|M.abscessus_ATCC_1997 YALLIGAGAVTLSRWRPARPVARGVVYGFIGTVTVASVAMAVIRLPIVSE
MLBr_01389|M.leprae_Br4923 LILTAPAAAIVIAVCIVTVARKPWPIAGVLVLFAVAAFPNYLFTQRGRYA
MAV_3138|M.avium_104 LIFTAPAMAIVLAVCIVTLARRPWPIAGAVLLCAVAALPNYLFVQRWPYA
Mflv_2269|M.gilvum_PYR-GCK APGIAALIGISVVTLWRGRGLFAPRLILAAMSAVTGVWAFVLLDRTPEWQ
.: : : .. . ::
MSMEG_3546|M.smegmatis_MC2_155 --------------ITPPDVGDR----IAKAAATAYRDLPEDERDRTVLL
MAB_1277|M.abscessus_ATCC_1997 AAAARWFAWAGGMGPTMLAEGDHSTEAFRQVARDTYDSLPADVRHRTALV
MLBr_01389|M.leprae_Br4923 KEG-------WDYSQVADVISSQAAPGDCLIVDNTVPWRPGPIRALLAAR
MAV_3138|M.avium_104 KEG-------WDYSQVADLIGSHAAPGDCLLVDNTVPWRPGPIRALLATR
Mflv_2269|M.gilvum_PYR-GCK PWLR-WVVLVGSIAVAVVFSAGAHRLGRATVAVAAAAIVLGAAAPLAYAL
. .. . :
MSMEG_3546|M.smegmatis_MC2_155 GESYITAAYLDGAAPRYGLPQAYGTNRS-----------YGYFTPPPDTA
MAB_1277|M.abscessus_ATCC_1997 VQIYPMAAAYDVEAGRAGISRAYSFHRG-----------YYYFGAPPQSM
MLBr_01389|M.leprae_Br4923 PAAFRSLIDIERGFYGPTVGTLWDGHVP-----------VWLVTAKINKC
MAV_3138|M.avium_104 PAAFRSLIDVERGAYGPKVGTLWDGHVA-----------IWLTTAKINKC
Mflv_2269|M.gilvum_PYR-GCK ETALHSHAGGAMAMSGPARDSGFGGPGGPAGPDGDNTELQELVRSADNRW
:. . :
MSMEG_3546|M.smegmatis_MC2_155 DEALYVGAE-PD-GMREHFVTVRNLATIDG----DLKLFLLRERRHPWPQ
MAB_1277|M.abscessus_ATCC_1997 TDMMYVGVDDPDPKLAQGFRGVQRIELLHAAGEDEAHVYRYYGRIAPWQQ
MLBr_01389|M.leprae_Br4923 STVWTVSDRDTSLPDHQAGQLLSPGLILGRAPAYQFPSYLGFRIVERWQF
MAV_3138|M.avium_104 STIWTITNKDNSLPDHQSGQSLPPGSAFGQAPAYRFPGYLGFHIVERWQF
Mflv_2269|M.gilvum_PYR-GCK AAAGVGSMLTGDLQLRTGASVMAIGGFTGGDNAPTLEQFQQYVADGQVRY
. : :
MSMEG_3546|M.smegmatis_MC2_155 IWSEQRSLTVS-------------------------------
MAB_1277|M.abscessus_ATCC_1997 LWDDWRTYK---------------------------------
MLBr_01389|M.leprae_Br4923 HYSQVIKSTR--------------------------------
MAV_3138|M.avium_104 HYSQVVKSTR--------------------------------
Mflv_2269|M.gilvum_PYR-GCK FIAGGHGGPGGGARSASQITAWVTQTFAPRDVGGTAVYDLQR