For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
HTRGRRTPGAAVQSSAVSDAAHVGDIVGAFGRIRRDGGGADGAAGGRWRRLRTLAVITGPGLIVMVGDND AGGVATYAQAGQNYGMGLLWTLVLLIPVLYVNQEMVLRLGAVARVGHARLIFERFGRFWGAFSVGDLLIL NALTIVTEFIGVALALGFLGCPKIVAVPAAAALLFAVVAGGSFRRWERLMFLLIAVNVLIFPMVLLVHPA PKTTVAGLIPQFPGGLNSTVLLLVVAIVGTTVAPWQLFFQQSNVVDKRITARWIPYGRADLVIGIVVVMV GATALMAVTAFGLAGTAAAGHFTDAGAVAAGLSAHLGRTVGVLFAIILLDASLIGANAVGLATSYAVGDA MGKRHSLHWKITEAPLFYGGYAVLLAVSAAVSFSPDHILGLVTQGVQALAGVLLPSATVFLVLLCNDRAV LGPWVNTVRQNIAAWTIVWCLVLLSLALTATTFFPDLSTGTIEAGLAAGAVLGVVAGAVMIVVGRRQRDL AEAEAIVRTLGGGLDPEQVDELDDASSLTRAERRAVRRQDRENWQTPSLALLDRPAMSPMRRAGLFTLRG YLVVAVVFVIIKLVQAGVVGPAASL*
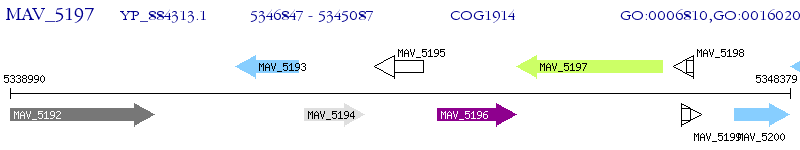
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5197 | - | - | 100% (586) | manganese/iron transporter |
| M. avium 104 | MAV_2120 | - | e-148 | 51.66% (573) | Mn2+/Fe2+ transporter |
| M. avium 104 | MAV_4571 | - | 3e-48 | 32.27% (406) | metal ion transporter, NRAMP family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0948c | mntH | 2e-18 | 25.36% (414) | putative manganese transport protein MntH |
| M. gilvum PYR-GCK | Mflv_1822 | - | 3e-17 | 26.71% (423) | Mn2+/Fe2+ transporter |
| M. tuberculosis H37Rv | Rv0924c | mntH | 2e-18 | 25.36% (414) | manganese transport protein MntH |
| M. leprae Br4923 | MLBr_02667 | - | 0.0 | 69.15% (577) | putative membrane transport protein |
| M. abscessus ATCC 19977 | MAB_4716c | - | 0.0 | 66.85% (558) | membrane transport protein |
| M. marinum M | MMAR_0315 | - | 0.0 | 78.41% (579) | divalent cation-transport integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_5589 | - | 5e-17 | 26.95% (423) | manganese transport protein MntH |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4807 | - | 0.0 | 78.41% (579) | divalent cation-transport integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_0718 | - | 3e-07 | 25.72% (311) | amino acid permease-associated region |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0315|M.marinum_M --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 --------------------------------------------------
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 MTTTSDQNAAAPPRFDGLRALFINATLKRSPELSHTDGLIERSSGIMREH
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0315|M.marinum_M --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 --------------------------------------------------
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 GVQVDTLRAVDHDIATGVWPDMTEHGWATDEWPALYRRVLDAHILVLCGP
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0315|M.marinum_M --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 --------------------------------------------------
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 IWLGDNSSVMKRVIERLYACSSLLNEDGQYAYYGRAGGCLITGNEDGVKH
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0315|M.marinum_M --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 ------------------------------------------MSLIIFEH
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 CAMNVLYSLQHLGYTIPPQADAGWIGEAGPGPSYLDPGSGGPENDFTNRN
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0315|M.marinum_M -------MLMTSISTSRR-------------------GAPNSAVHSADAA
MUL_4807|M.ulcerans_Agy99 ---------MTSISTSRR-------------------GAPNSAVHSADAA
MAV_5197|M.avium_104 ------------MHTRGR-------------------RTPGAAVQSS--A
MLBr_02667|M.leprae_Br4923 EPVDNSENPMTYISTDAISAVAVPPTTGDAIMCVKNCATPRVTPDTSGSS
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 TTFMTFNLMHIAQMLRAPAASRPTAINAPSGTPAAGRTSPTPTTDSQRRS
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0315|M.marinum_M VLDSAHTGDIVGAFGRIRRDG---DGVASRRWQRLRTLMVISGPGLIVMV
MUL_4807|M.ulcerans_Agy99 VLDSAHTGDIVGAFGRIRRDG---DGVASRRWQRLRTLMVISGPGLIVMV
MAV_5197|M.avium_104 VSDAAHVGDIVGAFGRIRRDGGGADGAAGGRWRRLRTLAVITGPGLIVMV
MLBr_02667|M.leprae_Br4923 VLDSAHVGDIVGAFGRIGWDV---THASRSRWQQLRLMMVITGPGLIAMI
MAB_4716c|M.abscessus_ATCC_199 -MDSAHVGDIVGALGTIGQHE---NIEGRSRWQRFRMLLAVLGPGLIVMV
Mb0948c|M.bovis_AF2122/97 RVARWLVAGEFRLLSHLCSRGSKVGELAQDTRTSLKTSWYLLGPAFVAAI
Rv0924c|M.tuberculosis_H37Rv ------MAGEFRLLSHLCSRGSKVGELAQDTRTSLKTSWYLLGPAFVAAI
Mflv_1822|M.gilvum_PYR-GCK --------------------------MTGDVRPQAKR-WLLLGPAFVAAI
MSMEG_5589|M.smegmatis_MC2_155 --------------------------MLEDTKVRARPSWLLLGPAFVAAI
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------MSSPGGSTLDRRLGVFDTVTIGLG
: .. :
MMAR_0315|M.marinum_M GDNDAGGVATYAQAGQNYGMTLLWTLALLIPVLYVNQEMVLRLGAVVRVG
MUL_4807|M.ulcerans_Agy99 GDNDAGGVATYAQAGQNYGMTLLWTLALLIPVLYVNQEMVLRLGAVVRVG
MAV_5197|M.avium_104 GDNDAGGVATYAQAGQNYGMGLLWTLVLLIPVLYVNQEMVLRLGAVARVG
MLBr_02667|M.leprae_Br4923 GDNDAGGVATYAQAGQKYGMNLLWTLVLLIPVLYVNQEMVLRLGAVTRVG
MAB_4716c|M.abscessus_ATCC_199 GDNDAGGVATYAQAGQNYGMALLWTLALLIPVLYVNQEMVVRLGAVAGVG
Mb0948c|M.bovis_AF2122/97 AYVDPGNVAANVSSGAQFGYLLLWVIVAANVMAALVQYLSAKLGLVTGRS
Rv0924c|M.tuberculosis_H37Rv AYVDPGNVAANVSSGAQFGYLLLWVIVAANVMAALVQYLSAKLGLVTGRS
Mflv_1822|M.gilvum_PYR-GCK AYVDPGNVAANVSAGAQFGFLLVWVIVAANLMACLVQYLSAKLGLVTGRS
MSMEG_5589|M.smegmatis_MC2_155 AYVDPGNVAANVSAGAQYGFLLVWVIVVANVMAGLVQYLSAKLGLVTGRS
Mvan_0718|M.vanbaalenii_PYR-1 SMIGAGIFVALAPAAAAAGSGLLIGLAVAAVVAVCNAMSSARLAARYPQS
. ..* ..: . :. * *: :. : :*. .
MMAR_0315|M.marinum_M HARLIF--ERFGKFWGAFSVGDLLILNALTIVTEFIGVSMALGFLG----
MUL_4807|M.ulcerans_Agy99 HARLIF--ERFGKFWGAFSVGDLLILNALTIVTEFIGVSMALGFLG----
MAV_5197|M.avium_104 HARLIF--ERFGRFWGAFSVGDLLILNALTIVTEFIGVALALGFLG----
MLBr_02667|M.leprae_Br4923 HARLIF--ERFGKFWGAFSVGDLLILNALTIVTEFIGVSLALDFLG----
MAB_4716c|M.abscessus_ATCC_199 HARLIF--SRFGRFWGAFSVGDLFVVNALTIVTEFIGVAMALSYFG----
Mb0948c|M.bovis_AF2122/97 LPEAIG--KRMGRPARLAYWAQAEIVAMATDVAEVIGGAIALRIMFNLPL
Rv0924c|M.tuberculosis_H37Rv LPEAIG--KRMGRPARLAYWAQAEIVAMATDVAEVIGGAIALRIMFNLPL
Mflv_1822|M.gilvum_PYR-GCK LPEAVG--SRMSRRTRLAYWVQAELVAMATDLAEVVGGAIALYLLFDLPL
MSMEG_5589|M.smegmatis_MC2_155 LPEAVA--DHTRTPTRVAFWLQAELVAMATDLAEVVGGAIALYLLFDLPL
Mvan_0718|M.vanbaalenii_PYR-1 GGTYVYGRERLGPFWGHAAGWSFIVGKTASCAAMALTVGFYAWPEY----
: .: . : : : : .:
MMAR_0315|M.marinum_M CPKAVAIPAAAVLLFAVVAGGSFRRWEQMMF-LLIAVNVVIIPMALLVHP
MUL_4807|M.ulcerans_Agy99 CPKAVAIPAAAVLLFAVVAGGSFRRWEQMMF-LLIAVNVVIIPMALLVHP
MAV_5197|M.avium_104 CPKIVAVPAAAALLFAVVAGGSFRRWERLMF-LLIAVNVLIFPMVLLVHP
MLBr_02667|M.leprae_Br4923 CPKTIAIPVAAVLLFAMVAGGSFRRWEQVMF-MLISVNVAVIPLVLSVHP
MAB_4716c|M.abscessus_ATCC_199 LPRWISVPLSAVLLFAVVAGGSFRRWERFLF-ALIAINIVMIPMAILVHP
Mb0948c|M.bovis_AF2122/97 PIGGIITGVVSLLLLTIQDRRGQRLFERVITALLLVIAIGFTASFFVVTP
Rv0924c|M.tuberculosis_H37Rv PIGGIITGVVSLLLLTIQDRRGQRLFERVITALLLVIAIGFTASFFVVTP
Mflv_1822|M.gilvum_PYR-GCK LVGGIITGAVSLILLMVKDLRGQRVFERVITGLLMVIAIGFLTSLFAASP
MSMEG_5589|M.smegmatis_MC2_155 LVGGLITGVVSLLLLAVQNRRGQRVFERVITGLLMVIAIGFLTSLFVKTP
Mvan_0718|M.vanbaalenii_PYR-1 ---ANAVAVASVVALTAVGYAGVQRSAALTR-IIVALVLAVLAMVVVVLL
: : : . : . :: : : . . .
MMAR_0315|M.marinum_M TFSGTVGGLVPRFPGGLDS---TVLLLIVAIVGTTIAPWQLFFQQSNVVD
MUL_4807|M.ulcerans_Agy99 TFSGTVGGLVPRFPGGLDS---TVLLLIVAIVGTTIAPWQLFFQQSNVVD
MAV_5197|M.avium_104 APKTTVAGLIPQFPGGLNS---TVLLLVVAIVGTTVAPWQLFFQQSNVVD
MLBr_02667|M.leprae_Br4923 ALGPSLHGLLPSLPGKPDSPTSVVLLVIMAIVGTTVAPWQLFFQQSSVVD
MAB_4716c|M.abscessus_ATCC_199 SVSKTASGFVPSFPGGLNA---ELLMLIVAIVGTTVAPWQLFFQQSNIVD
Mb0948c|M.bovis_AF2122/97 PPNAVLGGLAPRFQG------TESVLLAAAIMGATVMPHAVYLHSGLARD
Rv0924c|M.tuberculosis_H37Rv PPNAVLGGLAPRFQG------TESVLLAAAIMGATVMPHAVYLHSGLARD
Mflv_1822|M.gilvum_PYR-GCK PVGAAAEGLIPRFDG------AESVLLAAAMLGATVMPHAVYLHSGLARD
MSMEG_5589|M.smegmatis_MC2_155 PAAAVADGLIPRFDG------AESILLATAMLGATVMPHAVYLHSGLARD
Mvan_0718|M.vanbaalenii_PYR-1 GFGGADTSRLALGDDVTVYG----VLQAAGLLFFAFAGYARIATLGEEVR
. . . :: .:: :. .
MMAR_0315|M.marinum_M KR-------ITARWIPYARADLVLGIVVVIVGATALMAVTAFGLAGTSDV
MUL_4807|M.ulcerans_Agy99 KR-------ITARWIPYARADLVLGIVVVIVGATALMAVTAFGLAGTSDV
MAV_5197|M.avium_104 KR-------ITARWIPYGRADLVIGIVVVMVGATALMAVTAFGLAGTAAA
MLBr_02667|M.leprae_Br4923 KR-------ITPRWMRYGRADLVVGIVGVMAGAIALMSATAFGLAGTADS
MAB_4716c|M.abscessus_ATCC_199 KR-------LTPRWIRYERVDLAVGIVVVMIGAVCIMGAAAFGLAGTDAG
Mb0948c|M.bovis_AF2122/97 RHGHPDPGPQRRRLLRVTRWDVGLAMLIAGGVNAAMLLVAALNMRGRGDT
Rv0924c|M.tuberculosis_H37Rv RHGHPDPGPQRRRLLRVTRWDVGLAMLIAGGVNAAMLLVAALNMRGRGDT
Mflv_1822|M.gilvum_PYR-GCK RHGHPGPGEPRRALLRITRWDVGIAMLVAGAVNLSMLLVAATNLQGMADT
MSMEG_5589|M.smegmatis_MC2_155 RHGHAE-GPRLRMLLRVTRFDVGLAMLVAGAVNMAMLLVAATNLQGRGDT
Mvan_0718|M.vanbaalenii_PYR-1 DPAR-----TIPRAVALALVIALMVYAVVAVAVVSELGVAGLASATAPLS
: : . . : .:.
MMAR_0315|M.marinum_M GHFSDAGAVATSLTNHFGSTVGILFAVVLLDASLIGANAIGLATTYAVGD
MUL_4807|M.ulcerans_Agy99 GYFSDAGAVATSLTNHFGSTVGILFAVVLLDASLIGANAIGLATTYAVGD
MAV_5197|M.avium_104 GHFTDAGAVAAGLSAHLGRTVGVLFAIILLDASLIGANAVGLATSYAVGD
MLBr_02667|M.leprae_Br4923 GNFTDAGAVASHLSDHVGHPIGVLFAIILLDASLIGANVVGLATTYALGD
MAB_4716c|M.abscessus_ATCC_199 GDFTDAGAVARLLHEHVSPTVGALFAILLLDASLIGANAVGLATTYALGD
Mb0948c|M.bovis_AF2122/97 ASIEGA---YHAVHDTLGATIAVLFAVGLLASGLASSSVGAYAGAMIMQG
Rv0924c|M.tuberculosis_H37Rv ASIEGA---YHAVHDTLGATIAVLFAVGLLASGLASSSVGAYAGAMIMQG
Mflv_1822|M.gilvum_PYR-GCK DSIEGA---HAAVQNTLGDTVALLFAIGLLASGLASSSVGAYAGAMIMQG
MSMEG_5589|M.smegmatis_MC2_155 DSIEGA---HAAVSDALGPTVALLFAIGLLASGLASTSVGAYAGAMIMQG
Mvan_0718|M.vanbaalenii_PYR-1 DAVRAAGFPGLTPVVNVGAAIAALGALLALLLGVSRTTLAMARDRYLPAG
. * .. .:. * *: * .: :. .
MMAR_0315|M.marinum_M AMGKRHSLHWKISEAPLFYGGYAALLAISAAVAFSPDYVLGLVTQGVQAL
MUL_4807|M.ulcerans_Agy99 AMGKRHSLHWKISEAPLFYGGYAALLAISAAVAFSPDYLLGLVTQGVQAL
MAV_5197|M.avium_104 AMGKRHSLHWKITEAPLFYGGYAVLLAVSAAVSFSPDHILGLVTQGVQAL
MLBr_02667|M.leprae_Br4923 ALGKPHSLHWKITEAPLFYGSYTALLGISAVVAFSPGHILGLITQGVQAL
MAB_4716c|M.abscessus_ATCC_199 TFGKRHSLHWKPTEAPVFYLGYAMLLVVAAAVSFSPDPVLGVLTQGVQAL
Mb0948c|M.bovis_AF2122/97 LLHW---------SVPMLVRRLITLGPALAILTLGFDPTRTLVLS--QVV
Rv0924c|M.tuberculosis_H37Rv LLHW---------SVPMLVRRLITLGPALAILTLGFDPTRTLVLS--QVV
Mflv_1822|M.gilvum_PYR-GCK LLRR---------SVPLLARRLITLLPALVILAVGIHPSRALVLS--QVV
MSMEG_5589|M.smegmatis_MC2_155 LLKK---------SYPLLLRRLVTLVPALLVLAIGVDPSRALVLS--QVV
Mvan_0718|M.vanbaalenii_PYR-1 LAAVHP-----KHGTPHHAEVAVGLVVAVVAAFVDLRGAIGFSSFGVLLY
* * .. .
MMAR_0315|M.marinum_M AGILLPSATVFLVLLCNDHAVLGPWVNTLRQNIIAWTTVWCLVLLSLALT
MUL_4807|M.ulcerans_Agy99 AGILLPSATVFLVLLCNDHAVLGPWVNTLRQNIIAWTTVWCLVLLSLALT
MAV_5197|M.avium_104 AGVLLPSATVFLVLLCNDRAVLGPWVNTVRQNIAAWTIVWCLVLLSLALT
MLBr_02667|M.leprae_Br4923 AGVLLPSATVFLVLLCNDRPVLGPWVNTARQNVFAWMIVWSLVVLSLMLT
MAB_4716c|M.abscessus_ATCC_199 AGVLLPSATVFLVLLCNDKAVLGPWVNSTRQNIVAGVIVWVLVLLSLSLT
Mb0948c|M.bovis_AF2122/97 LSFGIPFAVLPLVKLTGSPAVMGGDTNHRATTWVGWVVAVMVSLLNVMLI
Rv0924c|M.tuberculosis_H37Rv LSFGIPFAVLPLVKLTGSPAVMGGDTNHRATTWVGWVVAVMVSLLNVMLI
Mflv_1822|M.gilvum_PYR-GCK LSFGIPFALIPLVRLTADRALMGADVNHRVTTAAGWVVAGLISLLNVVLI
MSMEG_5589|M.smegmatis_MC2_155 LSFGIPFALIPLVRLTSDKALMGGDANHRVTTAFGWGVAGLISVLNVVLI
Mvan_0718|M.vanbaalenii_PYR-1 YAIANASAWTLGARLIPAIGLVGCLVLAFTLPLTSVLAGAAVVLVGMIAY
.. . * . * ::* . . : ::.:
MMAR_0315|M.marinum_M VTTFFPRLSTAAIEIGLVAGAAIGMMGGAAVLITGRRRSDRRDAEAISQS
MUL_4807|M.ulcerans_Agy99 VTTFFPRLSTAAIEIGLVAGAAIGMMGGAAVLITGRRRSDRRDAEAISQS
MAV_5197|M.avium_104 ATTFFPDLSTGTIEAGLAAGAVLGVVAGAVMIVVGRRQRDLAEAEAIVRT
MLBr_02667|M.leprae_Br4923 VVTLFPNLSTAAMVGGLGAGTALGVVGAAAVIVAG-WLAERRGAGGIAQQ
MAB_4716c|M.abscessus_ATCC_199 AATFFPDLTTTQLKVFFVVGAGVGLLGGLFVSLAK-SRAEHAAASAVAAE
Mb0948c|M.bovis_AF2122/97 YLTVTG--------------------------------------------
Rv0924c|M.tuberculosis_H37Rv YLTVTG--------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK YLTLWG--------------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 YLTVNG--------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 TIGGTR--------------------------------------------
MMAR_0315|M.marinum_M LGGGLDPGEVDEINDAKLLSRAERRAIRQQDRAQWCTPNLTDLARPAMSQ
MUL_4807|M.ulcerans_Agy99 LGGGLDPGEVDEINDAKLLSRAERRAIRQQDRAQWCTPNLTDLARPAMSP
MAV_5197|M.avium_104 LGGGLDPEQVDELDDASSLTRAERRAVRRQDRENWQTPSLALLDRPAMSP
MLBr_02667|M.leprae_Br4923 P--SCSGSEQAQFDALARLTRAERRALCDERRANWTMPVLANLDRPVMSP
MAB_4716c|M.abscessus_ATCC_199 LG-GLDPDQVQDLDSMPR-DRVLRRDIQWQERESWRTPALETLERPAWSP
Mb0948c|M.bovis_AF2122/97 --------------------------------------------------
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_0315|M.marinum_M AHRVGLFTLRAYLVVAVVLVVVKVVQAGIG------
MUL_4807|M.ulcerans_Agy99 AHRLGLFTLRAYLVVAVVLVVVKVVQAGIG------
MAV_5197|M.avium_104 MRRAGLFTLRGYLVVAVVFVIIKLVQAGVVGPAASL
MLBr_02667|M.leprae_Br4923 VRRIGLLMLRGYLVLACVLVVVKVVQVGVG------
MAB_4716c|M.abscessus_ATCC_199 LRTAGMIALRGYLVVAVLLVGVKVAQTIIG------
Mb0948c|M.bovis_AF2122/97 ------------------------------------
Rv0924c|M.tuberculosis_H37Rv ------------------------------------
Mflv_1822|M.gilvum_PYR-GCK ------------------------------------
MSMEG_5589|M.smegmatis_MC2_155 ------------------------------------
Mvan_0718|M.vanbaalenii_PYR-1 ------------------------------------