For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSISTSRRGAPNSAVHSADAAVLDSAHTGDIVGAFGRIRRDGDGVASRRWQRLRTLMVISGPGLIVMVGD NDAGGVATYAQAGQNYGMTLLWTLALLIPVLYVNQEMVLRLGAVVRVGHARLIFERFGKFWGAFSVGDLL ILNALTIVTEFIGVSMALGFLGCPKAVAIPAAAVLLFAVVAGGSFRRWEQMMFLLIAVNVVIIPMALLVH PTFSGTVGGLVPRFPGGLDSTVLLLIVAIVGTTIAPWQLFFQQSNVVDKRITARWIPYARADLVLGIVVV IVGATALMAVTAFGLAGTSDVGYFSDAGAVATSLTNHFGSTVGILFAVVLLDASLIGANAIGLATTYAVG DAMGKRHSLHWKISEAPLFYGGYAALLAISAAVAFSPDYLLGLVTQGVQALAGILLPSATVFLVLLCNDH AVLGPWVNTLRQNIIAWTTVWCLVLLSLALTVTTFFPRLSTAAIEIGLVAGAAIGMMGGAAVLITGRRRS DRRDAEAISQSLGGGLDPGEVDEINDAKLLSRAERRAIRQQDRAQWCTPNLTDLARPAMSPAHRLGLFTL RAYLVVAVVLVVVKVVQAGIG*
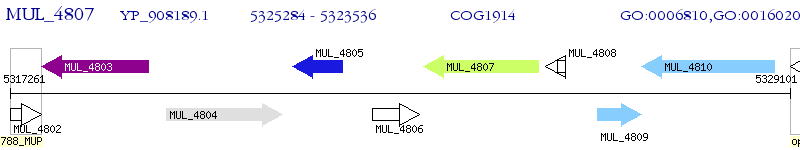
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4807 | - | - | 100% (582) | divalent cation-transport integral membrane protein |
| M. ulcerans Agy99 | MUL_3167 | - | 3e-49 | 32.41% (398) | divalent cation-transport integral membrane protein |
| M. ulcerans Agy99 | MUL_4445 | mntH | 3e-18 | 25.42% (417) | manganese transport protein MntH |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0948c | mntH | 1e-18 | 24.88% (414) | putative manganese transport protein MntH |
| M. gilvum PYR-GCK | Mflv_1822 | - | 2e-15 | 24.19% (434) | Mn2+/Fe2+ transporter |
| M. tuberculosis H37Rv | Rv0924c | mntH | 1e-18 | 24.88% (414) | manganese transport protein MntH |
| M. leprae Br4923 | MLBr_02667 | - | 0.0 | 68.52% (575) | putative membrane transport protein |
| M. abscessus ATCC 19977 | MAB_4716c | - | 0.0 | 65.47% (559) | membrane transport protein |
| M. marinum M | MMAR_0315 | - | 0.0 | 99.31% (582) | divalent cation-transport integral membrane protein |
| M. avium 104 | MAV_5197 | - | 0.0 | 78.41% (579) | manganese/iron transporter |
| M. smegmatis MC2 155 | MSMEG_1780 | - | 9e-17 | 22.25% (418) | hypothetical protein MSMEG_1780 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0315|M.marinum_M --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 --------------------------------------------------
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 MTTTSDQNAAAPPRFDGLRALFINATLKRSPELSHTDGLIERSSGIMREH
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1780|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0315|M.marinum_M --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 --------------------------------------------------
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 GVQVDTLRAVDHDIATGVWPDMTEHGWATDEWPALYRRVLDAHILVLCGP
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1780|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0315|M.marinum_M --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 --------------------------------------------------
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 IWLGDNSSVMKRVIERLYACSSLLNEDGQYAYYGRAGGCLITGNEDGVKH
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1780|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 --------------------------------------------------
MMAR_0315|M.marinum_M --------------------------------------------------
MAV_5197|M.avium_104 --------------------------------------------------
MLBr_02667|M.leprae_Br4923 ------------------------------------------MSLIIFEH
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 CAMNVLYSLQHLGYTIPPQADAGWIGEAGPGPSYLDPGSGGPENDFTNRN
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1780|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 ---------MTSISTSRR-------------------GAPNSAVHSADAA
MMAR_0315|M.marinum_M -------MLMTSISTSRR-------------------GAPNSAVHSADAA
MAV_5197|M.avium_104 ------------MHTRGR-------------------RTPGAAVQSS--A
MLBr_02667|M.leprae_Br4923 EPVDNSENPMTYISTDAISAVAVPPTTGDAIMCVKNCATPRVTPDTSGSS
MAB_4716c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0948c|M.bovis_AF2122/97 TTFMTFNLMHIAQMLRAPAASRPTAINAPSGTPAAGRTSPTPTTDSQRRS
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1780|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 VLDSAHTGDIVGAFGRIRRDG---DGVASRRWQRLRTLMVISGPGLIVMV
MMAR_0315|M.marinum_M VLDSAHTGDIVGAFGRIRRDG---DGVASRRWQRLRTLMVISGPGLIVMV
MAV_5197|M.avium_104 VSDAAHVGDIVGAFGRIRRDGGGADGAAGGRWRRLRTLAVITGPGLIVMV
MLBr_02667|M.leprae_Br4923 VLDSAHVGDIVGAFGRIGWDV---THASRSRWQQLRLMMVITGPGLIAMI
MAB_4716c|M.abscessus_ATCC_199 -MDSAHVGDIVGALGTIGQHE---NIEGRSRWQRFRMLLAVLGPGLIVMV
Mb0948c|M.bovis_AF2122/97 RVARWLVAGEFRLLSHLCSRGSKVGELAQDTRTSLKTSWYLLGPAFVAAI
Rv0924c|M.tuberculosis_H37Rv ------MAGEFRLLSHLCSRGSKVGELAQDTRTSLKTSWYLLGPAFVAAI
Mflv_1822|M.gilvum_PYR-GCK --------------------------MTGDVRPQAKR-WLLLGPAFVAAI
MSMEG_1780|M.smegmatis_MC2_155 ----------------------------------MKKVFAVALGILTAIG
: : : .
MUL_4807|M.ulcerans_Agy99 GDNDAGGVATYAQAGQNYGMTLLWTLALLIPVLYVNQEMVLRLGAVVRVG
MMAR_0315|M.marinum_M GDNDAGGVATYAQAGQNYGMTLLWTLALLIPVLYVNQEMVLRLGAVVRVG
MAV_5197|M.avium_104 GDNDAGGVATYAQAGQNYGMGLLWTLVLLIPVLYVNQEMVLRLGAVARVG
MLBr_02667|M.leprae_Br4923 GDNDAGGVATYAQAGQKYGMNLLWTLVLLIPVLYVNQEMVLRLGAVTRVG
MAB_4716c|M.abscessus_ATCC_199 GDNDAGGVATYAQAGQNYGMALLWTLALLIPVLYVNQEMVVRLGAVAGVG
Mb0948c|M.bovis_AF2122/97 AYVDPGNVAANVSSGAQFGYLLLWVIVAANVMAALVQYLSAKLGLVTGRS
Rv0924c|M.tuberculosis_H37Rv AYVDPGNVAANVSSGAQFGYLLLWVIVAANVMAALVQYLSAKLGLVTGRS
Mflv_1822|M.gilvum_PYR-GCK AYVDPGNVAANVSAGAQFGFLLVWVIVAANLMACLVQYLSAKLGLVTGRS
MSMEG_1780|M.smegmatis_MC2_155 GFLDIGDLVTNAVVGSRFGLALVWVVVVGVVGICLFAQMAGRVAAVSGRA
. * *.:.: . * .:* *:*.:. : : ::. * .
MUL_4807|M.ulcerans_Agy99 HARLIFERFGKFWGAFSVGDLLILNALTIVTEFIGVSMALGFLG----CP
MMAR_0315|M.marinum_M HARLIFERFGKFWGAFSVGDLLILNALTIVTEFIGVSMALGFLG----CP
MAV_5197|M.avium_104 HARLIFERFGRFWGAFSVGDLLILNALTIVTEFIGVALALGFLG----CP
MLBr_02667|M.leprae_Br4923 HARLIFERFGKFWGAFSVGDLLILNALTIVTEFIGVSLALDFLG----CP
MAB_4716c|M.abscessus_ATCC_199 HARLIFSRFGRFWGAFSVGDLFVVNALTIVTEFIGVAMALSYFG----LP
Mb0948c|M.bovis_AF2122/97 LPEAIGKRMGRPARLAYWAQAEIVAMATDVAEVIGGAIALRIMFNLPLPI
Rv0924c|M.tuberculosis_H37Rv LPEAIGKRMGRPARLAYWAQAEIVAMATDVAEVIGGAIALRIMFNLPLPI
Mflv_1822|M.gilvum_PYR-GCK LPEAVGSRMSRRTRLAYWVQAELVAMATDLAEVVGGAIALYLLFDLPLLV
MSMEG_1780|M.smegmatis_MC2_155 TFEIIRERLGPRTAAANLGASFFINLMTLTAEIGGIALALQLATD---VG
. : .*:. .: * :*. * ::**
MUL_4807|M.ulcerans_Agy99 KAVAIPAAAVLLFAVVAGGSFRRWEQMMF-LLIAVNVVIIPMALLVHPTF
MMAR_0315|M.marinum_M KAVAIPAAAVLLFAVVAGGSFRRWEQMMF-LLIAVNVVIIPMALLVHPTF
MAV_5197|M.avium_104 KIVAVPAAAALLFAVVAGGSFRRWERLMF-LLIAVNVLIFPMVLLVHPAP
MLBr_02667|M.leprae_Br4923 KTIAIPVAAVLLFAMVAGGSFRRWEQVMF-MLISVNVAVIPLVLSVHPAL
MAB_4716c|M.abscessus_ATCC_199 RWISVPLSAVLLFAVVAGGSFRRWERFLF-ALIAINIVMIPMAILVHPSV
Mb0948c|M.bovis_AF2122/97 GGIITGVVSLLLLTIQDRRGQRLFERVITALLLVIAIGFTASFFVVTPPP
Rv0924c|M.tuberculosis_H37Rv GGIITGVVSLLLLTIQDRRGQRLFERVITALLLVIAIGFTASFFVVTPPP
Mflv_1822|M.gilvum_PYR-GCK GGIITGAVSLILLMVKDLRGQRVFERVITGLLMVIAIGFLTSLFAASPPV
MSMEG_1780|M.smegmatis_MC2_155 PMMWIPVAAFAVWLVIWRVKFSVMENATG-ILGLCLIVFAVSVFALQPNW
: : : : *. * : . : *
MUL_4807|M.ulcerans_Agy99 SGTVGGLVPRFPGGLDS---TVLLLIVAIVGTTIAPWQLFFQQSNVVDKR
MMAR_0315|M.marinum_M SGTVGGLVPRFPGGLDS---TVLLLIVAIVGTTIAPWQLFFQQSNVVDKR
MAV_5197|M.avium_104 KTTVAGLIPQFPGGLNS---TVLLLVVAIVGTTVAPWQLFFQQSNVVDKR
MLBr_02667|M.leprae_Br4923 GPSLHGLLPSLPGKPDSPTSVVLLVIMAIVGTTVAPWQLFFQQSSVVDKR
MAB_4716c|M.abscessus_ATCC_199 SKTASGFVPSFPGGLNA---ELLMLIVAIVGTTVAPWQLFFQQSNIVDKR
Mb0948c|M.bovis_AF2122/97 NAVLGGLAPRFQG------TESVLLAAAIMGATVMPHAVYLHSGLARDRH
Rv0924c|M.tuberculosis_H37Rv NAVLGGLAPRFQG------TESVLLAAAIMGATVMPHAVYLHSGLARDRH
Mflv_1822|M.gilvum_PYR-GCK GAAAEGLIPRFDG------AESVLLAAAMLGATVMPHAVYLHSGLARDRH
MSMEG_1780|M.smegmatis_MC2_155 GDLAGQAVQPAIPESES-AATYWYYAIALFGAAMTPYEVFFFSSGAVEEH
*:.*::: * ::: .. :.:
MUL_4807|M.ulcerans_Agy99 -------ITARWIPYARADLVLGIVVVIVGATALMAVTAFGLAGTSDVGY
MMAR_0315|M.marinum_M -------ITARWIPYARADLVLGIVVVIVGATALMAVTAFGLAGTSDVGH
MAV_5197|M.avium_104 -------ITARWIPYGRADLVIGIVVVMVGATALMAVTAFGLAGTAAAGH
MLBr_02667|M.leprae_Br4923 -------ITPRWMRYGRADLVVGIVGVMAGAIALMSATAFGLAGTADSGN
MAB_4716c|M.abscessus_ATCC_199 -------LTPRWIRYERVDLAVGIVVVMIGAVCIMGAAAFGLAGTDAGGD
Mb0948c|M.bovis_AF2122/97 GHPDPGPQRRRLLRVTRWDVGLAMLIAGGVNAAMLLVAALNMRGRGDTAS
Rv0924c|M.tuberculosis_H37Rv GHPDPGPQRRRLLRVTRWDVGLAMLIAGGVNAAMLLVAALNMRGRGDTAS
Mflv_1822|M.gilvum_PYR-GCK GHPGPGEPRRALLRITRWDVGIAMLVAGAVNLSMLLVAATNLQGMADTDS
MSMEG_1780|M.smegmatis_MC2_155 -------WTTKDLGVSRMNVMVGFPLGGLLSIAIATCATLVLLP--QQVE
: * :: :.: .: :: :
MUL_4807|M.ulcerans_Agy99 FSDAGAVATSLTNHFGSTVGILFAVVLLDASLIGANAIGLATTYAVGDAM
MMAR_0315|M.marinum_M FSDAGAVATSLTNHFGSTVGILFAVVLLDASLIGANAIGLATTYAVGDAM
MAV_5197|M.avium_104 FTDAGAVAAGLSAHLGRTVGVLFAIILLDASLIGANAVGLATSYAVGDAM
MLBr_02667|M.leprae_Br4923 FTDAGAVASHLSDHVGHPIGVLFAIILLDASLIGANVVGLATTYALGDAL
MAB_4716c|M.abscessus_ATCC_199 FTDAGAVARLLHEHVSPTVGALFAILLLDASLIGANAVGLATTYALGDTF
Mb0948c|M.bovis_AF2122/97 IEGA---YHAVHDTLGATIAVLFAVGLLASGLASS---------SVGAYA
Rv0924c|M.tuberculosis_H37Rv IEGA---YHAVHDTLGATIAVLFAVGLLASGLASS---------SVGAYA
Mflv_1822|M.gilvum_PYR-GCK IEGA---HAAVQNTLGDTVALLFAIGLLASGLASS---------SVGAYA
MSMEG_1780|M.smegmatis_MC2_155 VTSLSQVVMPVVEAGGKLALAFAVIGIVAATFGAALETTLSSGYTLAQFL
. . : . : .: :: : : .: ::.
MUL_4807|M.ulcerans_Agy99 GKRHSLHWKISEAPLFYGGYAALLAISAAVAFSPDYLLGLVTQGVQALAG
MMAR_0315|M.marinum_M GKRHSLHWKISEAPLFYGGYAALLAISAAVAFSPDYVLGLVTQGVQALAG
MAV_5197|M.avium_104 GKRHSLHWKITEAPLFYGGYAVLLAVSAAVSFSPDHILGLVTQGVQALAG
MLBr_02667|M.leprae_Br4923 GKPHSLHWKITEAPLFYGSYTALLGISAVVAFSPGHILGLITQGVQALAG
MAB_4716c|M.abscessus_ATCC_199 GKRHSLHWKPTEAPVFYLGYAMLLVVAAAVSFSPDPVLGVLTQGVQALAG
Mb0948c|M.bovis_AF2122/97 GAMIMQGLLHWSVPMLVRRLITLGPALAILTLGFDPTRTLVLS--QVVLS
Rv0924c|M.tuberculosis_H37Rv GAMIMQGLLHWSVPMLVRRLITLGPALAILTLGFDPTRTLVLS--QVVLS
Mflv_1822|M.gilvum_PYR-GCK GAMIMQGLLRRSVPLLARRLITLLPALVILAVGIHPSRALVLS--QVVLS
MSMEG_1780|M.smegmatis_MC2_155 GWPWGKFRRPAQAARFHVVMFVSIVVGAAVLFT-GVDPVLVTEYSVVFSA
* ... : . : . :: . .. .
MUL_4807|M.ulcerans_Agy99 ILLPSATVFLVLLCNDHAVLGPWVNTLRQNIIAWTTVWCLVLLSLALTVT
MMAR_0315|M.marinum_M ILLPSATVFLVLLCNDHAVLGPWVNTLRQNIIAWTTVWCLVLLSLALTVT
MAV_5197|M.avium_104 VLLPSATVFLVLLCNDRAVLGPWVNTVRQNIAAWTIVWCLVLLSLALTAT
MLBr_02667|M.leprae_Br4923 VLLPSATVFLVLLCNDRPVLGPWVNTARQNVFAWMIVWSLVVLSLMLTVV
MAB_4716c|M.abscessus_ATCC_199 VLLPSATVFLVLLCNDKAVLGPWVNSTRQNIVAGVIVWVLVLLSLSLTAA
Mb0948c|M.bovis_AF2122/97 FGIPFAVLPLVKLTGSPAVMGGDTNHRATTWVGWVVAVMVSLLNVMLIYL
Rv0924c|M.tuberculosis_H37Rv FGIPFAVLPLVKLTGSPAVMGGDTNHRATTWVGWVVAVMVSLLNVMLIYL
Mflv_1822|M.gilvum_PYR-GCK FGIPFALIPLVRLTADRALMGADVNHRVTTAAGWVVAGLISLLNVVLIYL
MSMEG_1780|M.smegmatis_MC2_155 IALPLTYLPILIVANDPEYMGENTNGKVTNMFGTIYLVIILAASLAAIPL
. :* : : :: : . :* .* . . : .:
MUL_4807|M.ulcerans_Agy99 TFFPRLSTAAIEIGLVAGAAIGMMGGAAVLITGRRRSDRRDAEAISQSLG
MMAR_0315|M.marinum_M TFFPRLSTAAIEIGLVAGAAIGMMGGAAVLITGRRRSDRRDAEAISQSLG
MAV_5197|M.avium_104 TFFPDLSTGTIEAGLAAGAVLGVVAGAVMIVVGRRQRDLAEAEAIVRTLG
MLBr_02667|M.leprae_Br4923 TLFPNLSTAAMVGGLGAGTALGVVGAAAVIVAG-WLAERRGAGGIAQQP-
MAB_4716c|M.abscessus_ATCC_199 TFFPDLTTTQLKVFFVVGAGVGLLGGLFVSLAK-SRAEHAAASAVAAELG
Mb0948c|M.bovis_AF2122/97 TVTG----------------------------------------------
Rv0924c|M.tuberculosis_H37Rv TVTG----------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK TLWG----------------------------------------------
MSMEG_1780|M.smegmatis_MC2_155 MIVTGAGQ------------------------------------------
.
MUL_4807|M.ulcerans_Agy99 GGLDPGEVDEINDAKLLSRAERRAIRQQDRAQWCTPNLTDLARPAMSPAH
MMAR_0315|M.marinum_M GGLDPGEVDEINDAKLLSRAERRAIRQQDRAQWCTPNLTDLARPAMSQAH
MAV_5197|M.avium_104 GGLDPEQVDELDDASSLTRAERRAVRRQDRENWQTPSLALLDRPAMSPMR
MLBr_02667|M.leprae_Br4923 -SCSGSEQAQFDALARLTRAERRALCDERRANWTMPVLANLDRPVMSPVR
MAB_4716c|M.abscessus_ATCC_199 -GLDPDQVQDLDSMPR-DRVLRRDIQWQERESWRTPALETLERPAWSPLR
Mb0948c|M.bovis_AF2122/97 --------------------------------------------------
Rv0924c|M.tuberculosis_H37Rv --------------------------------------------------
Mflv_1822|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1780|M.smegmatis_MC2_155 --------------------------------------------------
MUL_4807|M.ulcerans_Agy99 RLGLFTLRAYLVVAVVLVVVKVVQAGIG------
MMAR_0315|M.marinum_M RVGLFTLRAYLVVAVVLVVVKVVQAGIG------
MAV_5197|M.avium_104 RAGLFTLRGYLVVAVVFVIIKLVQAGVVGPAASL
MLBr_02667|M.leprae_Br4923 RIGLLMLRGYLVLACVLVVVKVVQVGVG------
MAB_4716c|M.abscessus_ATCC_199 TAGMIALRGYLVVAVLLVGVKVAQTIIG------
Mb0948c|M.bovis_AF2122/97 ----------------------------------
Rv0924c|M.tuberculosis_H37Rv ----------------------------------
Mflv_1822|M.gilvum_PYR-GCK ----------------------------------
MSMEG_1780|M.smegmatis_MC2_155 ----------------------------------