For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GRVDGKVALISGGARGMGAEHARLLAAEGAKVVIGDILDDEGKAVADEIGDSVRYVHLDVTQPDQWDAAV ETAVGEFGKLNVLVNNAGTVALGPLKSFDLAKWQKVIDVNLTGTFLGMRVAVEPMIAAGGGSIINISSIE GLRGAPMVHPYVASKWGVRGLAKSAALELAPHNIRVNSVHPGFIRTPMTKHLPDDMVTVPLGRPAESREV STFVLFLASDESSYATGSEFVMDGGLVTDVPHKQF*
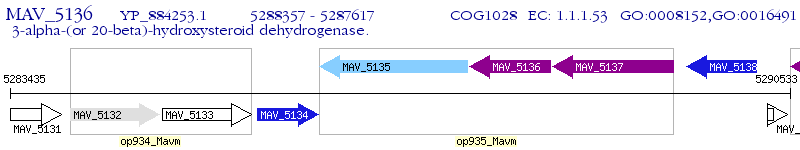
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5136 | - | - | 100% (246) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. avium 104 | MAV_2509 | - | 2e-89 | 63.52% (244) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. avium 104 | MAV_5088 | fabG | 1e-43 | 43.51% (239) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 4e-87 | 63.27% (245) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3281 | - | 1e-109 | 78.63% (248) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 4e-87 | 63.27% (245) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-30 | 37.34% (233) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 5e-56 | 45.38% (238) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0387 | fabG3 | 1e-122 | 86.99% (246) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. smegmatis MC2 155 | MSMEG_3515 | - | 1e-108 | 77.46% (244) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_3954 | - | 3e-44 | 43.40% (235) | PUTATIVE 3-oxoacyl-[acyl-carrier-protein] reductase |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 1e-122 | 86.59% (246) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | 1e-110 | 79.84% (248) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 --------------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVF
Rv2002|M.tuberculosis_H37Rv --------------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVF
MMAR_0387|M.marinum_M ---------------MGRVDGKVALISGGARGMGASHARLLVQEGAKVVI
MUL_1037|M.ulcerans_Agy99 ---------------MGRVDGKVALISGGARGMGASHARLLVQEGAKVVI
MAV_5136|M.avium_104 ---------------MGRVDGKVALISGGARGMGAEHARLLAAEGAKVVI
Mflv_3281|M.gilvum_PYR-GCK ---------------MGRVDGKVALISGGAQGMGAEDARALIAEGAKVVI
Mvan_3000|M.vanbaalenii_PYR-1 ---------------MGRVDGKVALISGGAQGMGAADARALIAEGAKVVI
MSMEG_3515|M.smegmatis_MC2_155 ---------------MGRVAGKVALISGGARGMGAAHARALVAEGAKVVI
MAB_4680c|M.abscessus_ATCC_199 -----------MNGNSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVI
TH_3954|M.thermoresistible__bu -------------MPSALLAEKVAVVTGAGRGIGREIARVLHAHGAEVVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
. : :::*. :*:* * : .* .*..
Mb2025|M.bovis_AF2122/97 GDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGL
Rv2002|M.tuberculosis_H37Rv GDILDEEGKAVAAELA----DAARYVHLDVTQPAQWTAAVDTAVTAFGGL
MMAR_0387|M.marinum_M GDILDEEGKALAEEIG----DAARYVHLDVTQPDQWEAAVATAVDEFGKL
MUL_1037|M.ulcerans_Agy99 GDILDEEGKALAEEIG----DAARYVHLDVTQPDQWEAAVATAVDEFGKL
MAV_5136|M.avium_104 GDILDDEGKAVADEIG----DSVRYVHLDVTQPDQWDAAVETAVGEFGKL
Mflv_3281|M.gilvum_PYR-GCK GDILDEKGQALADEINAQTPDSIRYVHLDVTQADQWEAAVATAVNDFGTL
Mvan_3000|M.vanbaalenii_PYR-1 GDILDDKGKALADEINAETPDSIRYVHLDVTQADQWEAAVATAVDAFGKL
MSMEG_3515|M.smegmatis_MC2_155 GDILDDEGAALAAELG----DAARYVHLDVTQAEDWDTAVATATTDFGLL
MAB_4680c|M.abscessus_ATCC_199 ADVLDGDGRQLAAELG----DAARFTHLDVTDAEQWRELIDTTLAGFGTI
TH_3954|M.thermoresistible__bu ADLDADAAAQAAAEVD------GTGVGCDVTAEDQMKRLVADTVAKHGRL
MLBr_01807|M.leprae_Br4923 THRASVVP------------DWFFGVECDVTDNDAIGAAFKAVEEHQGPV
. . *** . . * :
Mb2025|M.bovis_AF2122/97 HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGR
Rv2002|M.tuberculosis_H37Rv HVLVNNAGILNIGTIEDYALTEWQRILDVNLTGVFLGIRAVVKPMKEAGR
MMAR_0387|M.marinum_M DVLVNNAGIVALGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAAGS
MUL_1037|M.ulcerans_Agy99 DVLVNNVGIVALGQLKKFDLGKWQKVIDVNLTGTFLGMRAAVEPMTAAGS
MAV_5136|M.avium_104 NVLVNNAGTVALGPLKSFDLAKWQKVIDVNLTGTFLGMRVAVEPMIAAGG
Mflv_3281|M.gilvum_PYR-GCK NVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKAAGG
Mvan_3000|M.vanbaalenii_PYR-1 NVLVNNAGTVALGQIGQFDMAKWQKVIDVNLTGTFLGMQASVEAMKTAGG
MSMEG_3515|M.smegmatis_MC2_155 NVLINNAGIVALGAIGKFDMTKWQNVIDVNLTGTFLGMQASVGAMKAAGG
MAB_4680c|M.abscessus_ATCC_199 TGLVNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHGC
TH_3954|M.thermoresistible__bu DLFVNNAGITRDASLKRMTVADFDAVVTVHLRGTWLGVREATAVMREQKS
MLBr_01807|M.leprae_Br4923 EVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQRF
:: *.* : .: :: ::* *.: : *
Mb2025|M.bovis_AF2122/97 GSIINISSIEGLAGTVACHGYTATKFAVRGLTKSTALELGPSGIRVNSIH
Rv2002|M.tuberculosis_H37Rv GSIINISSIEGLAGTVACHGYTATKFAVRGLTKSTALELGPSGIRVNSIH
MMAR_0387|M.marinum_M GSIINVSSIEGLRGAPAVHPYVASKWAVRGLTKSAALELAPLNIRVNSIH
MUL_1037|M.ulcerans_Agy99 GSIINVSSIEGLRGAPAVHPYVASKWAVRGLTKSAALELAPLNIRVNSIH
MAV_5136|M.avium_104 GSIINISSIEGLRGAPMVHPYVASKWGVRGLAKSAALELAPHNIRVNSVH
Mflv_3281|M.gilvum_PYR-GCK GSIINISSIEGLRGAIMVHPYVASKWAVRGLTKSAALELGQYNIRVNSVH
Mvan_3000|M.vanbaalenii_PYR-1 GSIINISSIEGLRGAVMVHPYVASKWAVRGLTKSAALELGSHNIRVNSVH
MSMEG_3515|M.smegmatis_MC2_155 GSIINVSSIEGLRGAPMVHPYVASKWAVRGLTKSAALELGAHQIRVNSIH
MAB_4680c|M.abscessus_ATCC_199 GSIVNISSAAGLVGLALTSGYGASKWGVRGLSKVAAIELGRERIRVNSVH
TH_3954|M.thermoresistible__bu GSIVNISSLSGKSGNPGQTNYSAAKAGIIGLTKAAAKEVAHHNVRVNAIQ
MLBr_01807|M.leprae_Br4923 GRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVVA
* : :.* * * * *:* .: *::: * *:. : .* :
Mb2025|M.bovis_AF2122/97 PGLVKTPMTDWV-----PEDIFQTALGRAAEPVEVSNLVVYLASDESSYS
Rv2002|M.tuberculosis_H37Rv PGLVKTPMTDWV-----PEDIFQTALGRAAEPVEVSNLVVYLASDESSYS
MMAR_0387|M.marinum_M PGFIRTPMTANL-----PDDMVTIPLGRPAESREVSTFVVFLASDDASYA
MUL_1037|M.ulcerans_Agy99 PGFIRTPMTANL-----PDDMVTIPLGRPAESREVSTFVVFLASDDASYA
MAV_5136|M.avium_104 PGFIRTPMTKHL-----PDDMVTVPLGRPAESREVSTFVLFLASDESSYA
Mflv_3281|M.gilvum_PYR-GCK PGFIRTPMTKHF-----PDNMLRIPLGRPGQPEEVATFVVFLASDESRYS
Mvan_3000|M.vanbaalenii_PYR-1 PGFIRTPMTKHF-----PDNMLRIPLGRPGQPEEVATFVVFLASDESRYA
MSMEG_3515|M.smegmatis_MC2_155 PGFIRTPMTEHF-----PDDMLRIPLGRPGEVDEVSSFVVFLASDESRYA
MAB_4680c|M.abscessus_ATCC_199 PGVIYTPMTAKEGFREGEGNMPIAAMGRVGNADEVAGAVAYLLSDAASYV
TH_3954|M.thermoresistible__bu PGLIRTPMTETMPAEVFAEREAAVPMKRAGEPAEVAGAVVFLGSDLSSYI
MLBr_01807|M.leprae_Br4923 PGFIDTEMTRAMDSKYQEAALQAIPLGRIGAVEEIAGAVSFLASEDAGYI
**.: * ** .: * . *:: * :* *: : *
Mb2025|M.bovis_AF2122/97 TGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv TGAEFVVDGGTVAGLAHNDFGAVEVSSQPEWVT
MMAR_0387|M.marinum_M TGSEFVMDGGLVTDVPHKQF-------------
MUL_1037|M.ulcerans_Agy99 TGSEFVMDGGLVTDVPHKQF-------------
MAV_5136|M.avium_104 TGSEFVMDGGLVTDVPHKQF-------------
Mflv_3281|M.gilvum_PYR-GCK TGAEFVMDGGLTNDVPHK---------------
Mvan_3000|M.vanbaalenii_PYR-1 TGAEFVMDGGLTNDVPHK---------------
MSMEG_3515|M.smegmatis_MC2_155 TGAEFVMDGGLVNDVPHKL--------------
MAB_4680c|M.abscessus_ATCC_199 TGAELAVDGGWTAGPTLTTITGQ----------
TH_3954|M.thermoresistible__bu TGSVIEVGGGRYM--------------------
MLBr_01807|M.leprae_Br4923 SGAVIPVDGGLGMGH------------------
:*: : :.**