For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AHFQGKRYVVTGAASGIGNAVAERLLAAGAEVYCLDRNQPAANVTRHIEVDLANPHSIDAAVGQLDGDFD GLMNVAGIPGTAPADLVLAVNSLAVRHLTELFFDRLKPGGSVTIVSSTAGFGWPHRLDAIRDLLATDTYE EGAAWFKAHPQQGNAYNFSKEVSTVYTLSMGLAFAEMGFRINAVLPGPVQTPILPDFEESMGKDTLDGLK NLLGRHADPGDIADVVLFLASDDARWVNGQALAADGGISGAVASGVVPAPEI*
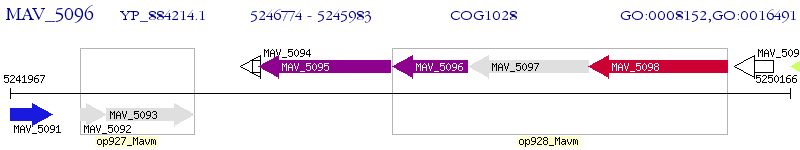
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5096 | - | - | 100% (263) | 3-alpha-hydroxysteroid dehydrogenase |
| M. avium 104 | MAV_0882 | - | 4e-45 | 40.64% (251) | 3-alpha-hydroxysteroid dehydrogenase |
| M. avium 104 | MAV_1264 | - | 2e-36 | 36.15% (260) | oxidoreductase, short chain dehydrogenase/reductase family |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0792 | - | 1e-16 | 29.24% (277) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4338 | - | 3e-45 | 39.00% (259) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0769 | - | 1e-16 | 29.24% (277) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01740 | - | 1e-10 | 28.45% (232) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0827 | - | 6e-16 | 29.15% (271) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4773 | - | 3e-42 | 37.65% (255) | dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_4862 | - | 2e-39 | 36.61% (254) | 3-alpha-hydroxysteroid dehydrogenase |
| M. thermoresistible (build 8) | TH_3914 | - | 1e-116 | 77.10% (262) | PUTATIVE 3-alpha-hydroxysteroid dehydrogenase |
| M. ulcerans Agy99 | MUL_0344 | - | 1e-44 | 38.82% (255) | 3-alpha-hydroxysteroid dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2008 | - | 9e-52 | 43.03% (251) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0792|M.bovis_AF2122/97 --------------------------------MFDSKVAIVTGAAQGIGQ
Rv0769|M.tuberculosis_H37Rv --------------------------------MFDSKVAIVTGAAQGIGQ
MAB_0827|M.abscessus_ATCC_1997 -----------------------------MTSPLKSRRAFVTGGSRGIGA
MAV_5096|M.avium_104 ------------------------------MAHFQGKRYVVTGAASGIGN
TH_3914|M.thermoresistible__bu ------------------------------MSEFTNRRYVVTGAASGIGD
Mflv_4338|M.gilvum_PYR-GCK MTRMGGKTVITTVEITRLVVPTMEGYQEGSAMSAEGRRVVVVGAGSGIGA
Mvan_2008|M.vanbaalenii_PYR-1 -------------------------------MQVEKRRIVVVGAGSGIGA
MMAR_4773|M.marinum_M ------------------------MAQIEQLWRYDGRRAVVTGCASGIGE
MUL_0344|M.ulcerans_Agy99 ------------------------MAQIEQLWRYDGRRAVVTGCASGIGE
MSMEG_4862|M.smegmatis_MC2_155 ------------------------MAALDALVRYDGRHVVVTGCSSGIGA
MLBr_01740|M.leprae_Br4923 --------------------------MVKTAQYFVGKRCFVTGAASGIGR
: .*.* . ***
Mb0792|M.bovis_AF2122/97 AYAQALAREGASVVVADIN-ADGAAAVAKQIVADGGTVIHVPVDVSDEDS
Rv0769|M.tuberculosis_H37Rv AYAQALAREGASVVVADIN-ADGAAAVAKQIVADGGTAIHVPVDVSDEDS
MAB_0827|M.abscessus_ATCC_1997 GIVRRLAEDGARVVFTYSSSVQQAADVVDDINEDGGWARAVKADVADPDQ
MAV_5096|M.avium_104 AVAERLLAAGAEVYCLDRNQP--AANVTRHIEVDLANPHSIDAAVGQLDG
TH_3914|M.thermoresistible__bu ATARKLLAAGAEVTSMDRNPP--SAPVSRHIEVDLANPRSIDAAVEQLEG
Mflv_4338|M.gilvum_PYR-GCK ATAAWFHSRGDHVLAVDARPR--SGPFAEWLVCDLRSADEIEGALGRIGH
Mvan_2008|M.vanbaalenii_PYR-1 AVAAHFHDNGDHVLAVDLRPN--DTPAAAHAQCDLRDPQSIAGLLDDIGA
MMAR_4773|M.marinum_M RAVHQLTGLGAQVVGLDQRQP--GYEISEFHQIDLADPGSIGRAVEAIGG
MUL_0344|M.ulcerans_Agy99 RAVHRLTGLGAQVVGLDQRQP--GYEISEFHQIDLADPGSIDRAVEAIGG
MSMEG_4862|M.smegmatis_MC2_155 KVTDQLRCLGARVTGLDLRAPDSGAGPDAFREMDLADPDSVDAAAAAIDG
MLBr_01740|M.leprae_Br4923 ATALRLAAYGAELYLTDRHCDGLAQTVSEARALGAQVPEYRALDISDYDE
. : * : .
Mb0792|M.bovis_AF2122/97 AKAMVDRAVGAFGGIDY--LVNNAAIYGGMKLDLLLTVPLDYYKKFMSVN
Rv0769|M.tuberculosis_H37Rv AKAMVDRAVGAFGGIDY--LVNNAAIYGGMKLDLLLTVPLDYYKKFMSVN
MAB_0827|M.abscessus_ATCC_1997 LGAAIGQAAEELGGLDI--LVNNAGILS---IGDVDTATLADFDEIVAVN
MAV_5096|M.avium_104 DFDGLMNVAGIP-GTAPADLVLAVNSLAVRHLTELFFDRLKPGGSVTIVS
TH_3914|M.thermoresistible__bu SYDGLMNIAGIP-GTAPGETVLAVNSLAVRHLTEAFLDRLNSGGTVTTVS
Mflv_4338|M.gilvum_PYR-GCK GWDVLAHVAGIP-GTADARDVLTVNYLGARLMLDGMLPLMNHGGSMVAVA
Mvan_2008|M.vanbaalenii_PYR-1 DWDVLAHVAGVP-GTAPADDVLTVNYLGMRKMVEGMLPRMRRGGSVVAVA
MMAR_4773|M.marinum_M PVDALFNIAGVSSGIGDPLLVVTINFLGLRQLTEALLPMMGPGSAIVSVS
MUL_0344|M.ulcerans_Agy99 PVDALFNIAGVSSGIGDPLLVVTINFLGLRHLTEALLPMMGPGSAIVSVS
MSMEG_4862|M.smegmatis_MC2_155 PVHALFNVAGVSSGIGDPLLVVRINFLGTRQFTEALVDRIADGGSITSVS
MLBr_01740|M.leprae_Br4923 VAAFGADIHARHPSMDIVMNIAGVSVWG-----TVDRLTHDQWSNVVAIN
. : . . :
Mb0792|M.bovis_AF2122/97 HDGVLVCTR--AVYKHMAKRGGGAIVN--QSSTAAWLYSNFYGLAKVGVN
Rv0769|M.tuberculosis_H37Rv HDGVLVCTR--AVYKHMAKRGGGAIVN--QSSTAAWLYSNFYGLAKVGVN
MAB_0827|M.abscessus_ATCC_1997 VRAVFAGIK--AASPHLTEGGRIVTIGSINGDSSPTANLSLYSMTKAAVA
MAV_5096|M.avium_104 STAGFGWPHRLDAIRDLLATDTYEEGAAWFKAHP--QQGNAYNFSKEVST
TH_3914|M.thermoresistible__bu STAGFGWPERLEAIRDLLATESFEDGARWFKEHP--QQGNAYNFSKEVTT
Mflv_4338|M.gilvum_PYR-GCK STAALGWDSNIGLLSGLLDARDTTAVTRWLEAQD--PSMAYYLS-KQALV
Mvan_2008|M.vanbaalenii_PYR-1 SIAGIGWEQRLDELSELLAATDPEAVAAWQSRQD--PSYPVYTTSKQAVI
MMAR_4773|M.marinum_M SLAAAGYREHHRAVAPLVNTTTMVEGIDWCKRHLEVLGAG-YQLSKEAVI
MUL_0344|M.ulcerans_Agy99 SLAAAGYREHHRAVAPLLNTTTMVEGIDWCKRHLEVLGAG-YQLSKEAVI
MSMEG_4862|M.smegmatis_MC2_155 SLAASAYRENRAVTAGLVSTGSVDEGLRWCAENPDALADGGYRLSKEAII
MLBr_01740|M.leprae_Br4923 LMGPIHIIETFVPAILAAGRGGHLVNVSSAAGLVALPWHAAYSASKYGLR
. * *
Mb0792|M.bovis_AF2122/97 GLTQQLA-RELGGMKIRINAIAPGPIDTEATRTVTPAELVKNMVQT-IPL
Rv0769|M.tuberculosis_H37Rv GLTQQLA-RELGGMKIRINAIAPGPIDTEATRTVTPAELVKNMVQT-IPL
MAB_0827|M.abscessus_ATCC_1997 GLTRGLA-REFGPRGITINNVQPGPIGTDMN--PEDGQLADILRPN-IAV
MAV_5096|M.avium_104 VYTLSMG-LAFAEMGFRINAVLPGPVQTPILPDFEESMGKDTLDGLKNLL
TH_3914|M.thermoresistible__bu VYTMTMG-LTAAQRGIRINAVLPGPVETPILADFEESMGKDTLDGVKALL
Mflv_4338|M.gilvum_PYR-GCK LYAKRLSGVAWREYGVRVNTVSPGPTETPILGDFEESMGRDMLDGVRTTI
Mvan_2008|M.vanbaalenii_PYR-1 LYAKRLAGTALSRYGVRVNTVSPGPVETPILADFEQTMGKEILDMLRDTY
MMAR_4773|M.marinum_M LYTMRSA-VGLGARGIRINCTGPGVTETPILDQLTTAYGAEFLDDIAKPL
MUL_0344|M.ulcerans_Agy99 LYTMRSA-VGLGARGIRINCTGPGVTETPILDQLTTAYGAEFPDDIAKPL
MSMEG_4862|M.smegmatis_MC2_155 LYGMSKA-VELGARGIRVNCTGPGVTDTPILDQLRSAYGQQYLDSFTTPL
MLBr_01740|M.leprae_Br4923 GLSEVLR-FDLARYRIGVSVVVPGAVRTPLVDTIEIAGVDRQDPRFSRWV
. :. ** *
Mb0792|M.bovis_AF2122/97 SRMG--------TPEDLVG---------------MCLFLLSDSASWITGQ
Rv0769|M.tuberculosis_H37Rv SRMG--------TPEDLVG---------------MCLFLLSDSASWITGQ
MAB_0827|M.abscessus_ATCC_1997 GRYG--------QPRDIAS---------------LVSYLVSDEAWYITGT
MAV_5096|M.avium_104 GRHA--------DPGDIAD---------------VVLFLASDDARWVNGQ
TH_3914|M.thermoresistible__bu GRHA--------SPDDIAD---------------VVVFLASDAARWINGQ
Mflv_4338|M.gilvum_PYR-GCK GRHG--------TVDDMTP---------------AIGFLSSPEARWINGQ
Mvan_2008|M.vanbaalenii_PYR-1 GRHG--------TVDDIVP---------------VIAFLASDAAQWINGQ
MMAR_4773|M.marinum_M GRVA--------TPGEQAS---------------VLVFLNGHAASYISGQ
MUL_0344|M.ulcerans_Agy99 GRVA--------TPGEQAS---------------VLVFLNGHSASYISGQ
MSMEG_4862|M.smegmatis_MC2_155 GRNS--------EPDEQAA---------------LLVFLGSAAASYVTGQ
MLBr_01740|M.leprae_Br4923 DRFRGHAVSAERAAEKILAGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGV
.* . : * : *
Mb0792|M.bovis_AF2122/97 IFNVDGGQIIRS--------------------------------
Rv0769|M.tuberculosis_H37Rv IFNVDGGQIIRS--------------------------------
MAB_0827|M.abscessus_ATCC_1997 AINIDGGFTA----------------------------------
MAV_5096|M.avium_104 ALAADGGISGAVASGVVPAPEI----------------------
TH_3914|M.thermoresistible__bu AVAVDGGITGAVASGVVPKPEF----------------------
Mflv_4338|M.gilvum_PYR-GCK DIHVDGGFTAALVHPMPERAQV----------------------
Mvan_2008|M.vanbaalenii_PYR-1 DVHVDSGFVTSMTAGTPIAL------------------------
MMAR_4773|M.marinum_M VVWVDGGNVAASIAGELGEGRA----------------------
MUL_0344|M.ulcerans_Agy99 VVRVDGGNVAASIAGELGEGRA----------------------
MSMEG_4862|M.smegmatis_MC2_155 VIWADGGIIAQRETALIESTRATQES------------------
MLBr_01740|M.leprae_Br4923 VMKQVNVVFTRALRPSPATMRCVEPIKVGVYSQPRSGEGGKSGN
. .