For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVSHLPADRVDETGARTVAQGIEDGVAAERLGFANVYLSERWNLKEAGVVLGAIGARTTRVGLGTGLVSP ARRHVLHMAALGATIHAAVGPRFLLGLGRGDHAYLRHEGLKTAGFAAVVDYVKIMRRLWDGEAVTYDGPA GNYRNIKLGDIYQGPAPQVLYGTFGMPKAAAAVAEAFDGVILPPMMSPSATTASVGRLRRACEAIGRDPA TLRVVQCVVTAPDLSVDETRALAHARAVTYLQAPEYGAALARVNGWSLAPVERLRAAVAKVKSAGDELVD TAFHRIQLMDLADMVPDEWMEASCAFGTAAQCARKLQEFKRAGADEIATYGSTPSQNAGVLAAWSDAFAG DMR
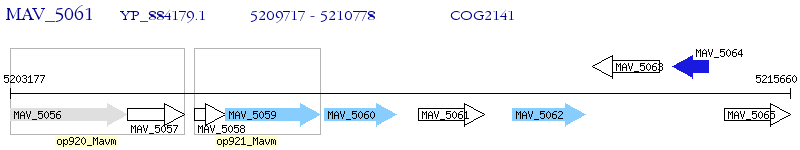
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5061 | - | - | 100% (353) | hypothetical protein MAV_5061 |
| M. avium 104 | MAV_1930 | - | 7e-58 | 38.17% (317) | hypothetical protein MAV_1930 |
| M. avium 104 | MAV_1607 | - | 2e-43 | 34.23% (333) | hypothetical protein MAV_1607 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3120c | - | 1e-12 | 26.46% (325) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_4328 | - | 5e-44 | 34.53% (333) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3093c | - | 5e-13 | 27.08% (325) | oxidoreductase |
| M. leprae Br4923 | MLBr_00269 | - | 5e-10 | 28.25% (177) | putative F420-dependent glucose-6-phosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2816c | - | 2e-17 | 27.96% (304) | putative luciferase-like protein |
| M. marinum M | MMAR_4003 | - | 4e-42 | 33.33% (333) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2249 | - | 5e-45 | 34.23% (333) | hypothetical protein MSMEG_2249 |
| M. thermoresistible (build 8) | TH_1627 | - | 3e-42 | 33.03% (333) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3866 | - | 5e-42 | 33.33% (333) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_2018 | - | 1e-43 | 34.23% (333) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3120c|M.bovis_AF2122/97 ----------------------------------MTDIEVALPFWLD-RP
Rv3093c|M.tuberculosis_H37Rv ----------------------------------MTDIEVALPFWLD-RP
MSMEG_2249|M.smegmatis_MC2_155 MNSRLFNSHRVVAGCVTECGIVVTNEEFDMSEHALDELGYYLLAGAGGEG
TH_1627|M.thermoresistible__bu MN------------------------------RPLDELGYYLLAGAGGEG
Mflv_4328|M.gilvum_PYR-GCK MTEA-----------------------------KMDELGYYLLAGAGGEG
Mvan_2018|M.vanbaalenii_PYR-1 MTR------------------------------VLDELGFYLLAGAGGEG
MMAR_4003|M.marinum_M MSDR-----------------------------TLNELGYYLLAGAGGEG
MUL_3866|M.ulcerans_Agy99 MSDR-----------------------------TLNELGYYLLAGAGGEG
MAV_5061|M.avium_104 ---------------------------------MVSHLPADRVDETGART
MAB_2816c|M.abscessus_ATCC_199 -------------------------------------MDFGVVLQCD-PP
MLBr_00269|M.leprae_Br4923 ----------------------------------MAELRLGYKASAEQFA
:
Mb3120c|M.bovis_AF2122/97 DHEATDVALAAADTGFAALW-IGE-MATYDAF-------ALATSIGLRTP
Rv3093c|M.tuberculosis_H37Rv DHEATDVALAAADTGFAALW-IGE-MATYDAF-------ALATSIGLRTP
MSMEG_2249|M.smegmatis_MC2_155 PATLMDEARRGEELGFGTAF-ISERWNVKEAS-------SLVGAACAVTT
TH_1627|M.thermoresistible__bu PATLMDEARRGEELGFGTAF-ISERWNVKEAS-------SLVGAACAVTE
Mflv_4328|M.gilvum_PYR-GCK PATLMDEARRGEELGFGTGF-ISERWNVKEAS-------SLTGAALAVTS
Mvan_2018|M.vanbaalenii_PYR-1 PATLMDEARRGEELGFGTGF-ISERWNVKEAS-------SLTGAALAVTS
MMAR_4003|M.marinum_M PATLMDEARRGEELGLGTAF-VSERWNVKEAS-------SLVGAACAVTS
MUL_3866|M.ulcerans_Agy99 PATLMDEARRGEELGLGTAF-VSERWNVKEAS-------SLVGAACAVTS
MAV_5061|M.avium_104 VAQGIEDGVAAERLGFANVY-LSERWNLKEAG-------VVLGAIGARTT
MAB_2816c|M.abscessus_ATCC_199 AVRTVQLAKLAEAHGFSHLWTFDSHLLWQEPY-------VLYSQILHETR
MLBr_00269|M.leprae_Br4923 PRELVELGVAAEAHGMDSATVSDHFQPWRHQGGHASFSLSWMTAVGERTN
: . . *: . . *
Mb3120c|M.bovis_AF2122/97 NMTLKVGPLAVGVRG-PVGLALGVSSVASLTGCRVDLALGASSPAIVAGW
Rv3093c|M.tuberculosis_H37Rv NMTLKVGPLAVGVRG-PVGLALGVSSVASLTGCRVDLALGASSPAIVAGW
MSMEG_2249|M.smegmatis_MC2_155 RMQIATAATNHNTRH-PLITGSWATTMHRLSGGRFTLGIGRGIAAMYGAF
TH_1627|M.thermoresistible__bu RMQIATAATNHNTRH-PLITGSWATTMHRLSGGRFTLGIGRGVAAMYAAF
Mflv_4328|M.gilvum_PYR-GCK RMQIATAATNHNTRH-PLITGSWATTMHRLSGGRFTLGIGRGIGAIYNAF
Mvan_2018|M.vanbaalenii_PYR-1 RMQIATAATNHNTRH-PLITGSWATTMHRLSGGRFTLGIGRGIGAIYNAF
MMAR_4003|M.marinum_M RMQIATAATNHNTRH-PLVTASWATTMHRLSGGRFTLGIGRGIAAIYGAF
MUL_3866|M.ulcerans_Agy99 RMQIATAATNHNTRH-PVVTASWATTMHRLSGGRFTLGIGRGIAAIYGAF
MAV_5061|M.avium_104 RVGLGTGLVSPARRH-VLHMAALGATIHAAVGPRFLLGLGRGDHAYLRHE
MAB_2816c|M.abscessus_ATCC_199 RIVVGPMVTNPATRD-WTVTASLYATLNDMYGNRTICGIGRGDSAVRVTG
MLBr_00269|M.leprae_Br4923 RILLGTSVLTPTFRYNPAVIGQAFATMGCLYPNRVFLGVGTGEALNEVAT
.: : * . ::: * .:* .
Mb3120c|M.bovis_AF2122/97 HGR----PWAHHVPVMRETIECLRSIFTGARVEYSGRHVNSRGFRLRG--
Rv3093c|M.tuberculosis_H37Rv HGR----PWAHHVPVMRETIECLRSIFTGARVEYSGRHVNSRGFRLRG--
MSMEG_2249|M.smegmatis_MC2_155 G------IPSVTTAQMEDFAQVMRRLWQGEVIFNHDGPIGNYQVLFLDPD
TH_1627|M.thermoresistible__bu G------IPPVTTAQMEDFAQVMRRLWRGEVIFDHDGPIGRYQVLFLDPD
Mflv_4328|M.gilvum_PYR-GCK G------IPTVTTAQMEDFAQVMRKLWHGELIFNHDGPLGKYQVLFLDPD
Mvan_2018|M.vanbaalenii_PYR-1 G------IPTVTTAQMEDFAQVMRKLWHGELIFNHDGPLGKYQVLFLDPD
MMAR_4003|M.marinum_M G------IPAVTTAQMEDWAQVMRRLWKGEMIFNHDGPMGRYPVLFLDPD
MUL_3866|M.ulcerans_Agy99 G------IPAVTTAQMEDWAQVMRRLWKGEMVFNHDGPMGRYPVLFLDPD
MAV_5061|M.avium_104 G------LKTAGFAAVVDYVKIMRRLWDGEAVT-YDGPAGNYRNIKLGDI
MAB_2816c|M.abscessus_ATCC_199 G-------TPTTLATLRESIHVIRELANSRPVEYHGATMQ------FPWS
MLBr_00269|M.leprae_Br4923 GYQGAWPEFKERFARLRESVRLMRELWRGDRVDFDGDYYQLKGASIYD-V
. : : . :* : . : .
Mb3120c|M.bovis_AF2122/97 AAPDTRIALGAFG--PGMIRLAAQHADEVVLN--LASPFRVGRVRAAIDS
Rv3093c|M.tuberculosis_H37Rv AAPDTRIALGAFG--PGMIRLAAQHADEVVLN--LASPFRVGRVRAAIDS
MSMEG_2249|M.smegmatis_MC2_155 FREDIRLAIVAFG--PQTLALGGREFDDVILHT-YFTPETLQRAVKTVKT
TH_1627|M.thermoresistible__bu FNEDIRLAIVAFG--PQTLALGGRMFDDVILHT-YFTPETLQRAVKTVKD
Mflv_4328|M.gilvum_PYR-GCK FREDIRLAIVAFG--PQTLALGGREFDDVILHT-YFTPETLQRAVKTVKD
Mvan_2018|M.vanbaalenii_PYR-1 FREDIRLAIVAFG--PQTLALGGREFDDVILHT-YFTPETLQRAVRTVKD
MMAR_4003|M.marinum_M FNEDIRLALVALG--PKSLALGGRVFDDVILHT-YFTPETLQRCVSIVKT
MUL_3866|M.ulcerans_Agy99 FNEDIRLALVALG--PKSLALGGRVFDDVILHT-YFTPETLQRCVSIVKT
MAV_5061|M.avium_104 YQGPAPQVLYGTFGMPKAAAAVAEAFDGVILPP-MMSPSATTASVGRLRR
MAB_2816c|M.abscessus_ATCC_199 RGSELDVWVAAYG--PKALALAGQVADGFILQ--LADIDIARWMIGAVRE
MLBr_00269|M.leprae_Br4923 PEGGVPIYIAAGG--PEVAKYAGRAGEGFVCTSGKGEELYTEKLIPAVLE
: . * .. : .: :
Mb3120c|M.bovis_AF2122/97 AAAAAGRAAPRLTVWVPVAVN----PGAAAHSQLAAQLAVYLAPPGYGEM
Rv3093c|M.tuberculosis_H37Rv AAAAAGRAAPRLTVCVPVAVN----PGAAAHSQLAAQLAVYLAPPGYGEM
MSMEG_2249|M.smegmatis_MC2_155 AAEQAGRDPGSVRVWSCFATVGDHLPEELRLKKTVARLATYLQG--YGDL
TH_1627|M.thermoresistible__bu AAEQAGRDPAEVRVWSCFATVGDHLPEELRLKKTVARLATYLQG--YGDL
Mflv_4328|M.gilvum_PYR-GCK AAEQAGRDPASVRVWSCFATVGDHLPEELRLKKTVARLATYLQG--YGDL
Mvan_2018|M.vanbaalenii_PYR-1 AAEQAGRDPASVRVWSCFATVGDHLPEELRLKKTVARLATYLQG--YGDL
MMAR_4003|M.marinum_M AADRAGRDPASVRVWSCFATVGDHLPEQLRLKKTVARLATYLQG--YGDL
MUL_3866|M.ulcerans_Agy99 AADRAGRDPASVRVWSCFATVGDHLPEQLRLKKTVARLATYLQG--YGDL
MAV_5061|M.avium_104 ACEAIGRDPATLRVVQCVVTAPDLSVDETR-ALAHARAVTYLQAPEYGAA
MAB_2816c|M.abscessus_ATCC_199 AAVAAGRDPAAVRICVAAPMYVG---SDRTHMRAQCRWFGGMVGNHVADI
MLBr_00269|M.leprae_Br4923 GAAVAGRDADDIDKMIEIKMSYD---PDPEQALSNIRFWAPLSL------
.. ** . : : :
Mb3120c|M.bovis_AF2122/97 FSALG------------------FDG--LVRSARSRATRRELAVAVPSEL
Rv3093c|M.tuberculosis_H37Rv FSALG------------------FDG--LVRSARSRATRRELAVAVPSEL
MSMEG_2249|M.smegmatis_MC2_155 LVNTNGWDPAVLQRFRDDKVVQSIPG--GIDHKATAEQIEHIATLIPDEW
TH_1627|M.thermoresistible__bu LVETNGWDPAVLRKFRDDPVVQSIPG--GIDHKATPEQIEHIATLIPDEW
Mflv_4328|M.gilvum_PYR-GCK LVNTNGWDPAVLQRFRDDKVVQSIGG--GIDHKATAEQIEHIATLIPDEW
Mvan_2018|M.vanbaalenii_PYR-1 LVNTNGWDPAVLQRFRDDKVVQSVGG--GIDHKATAEQIEHIATLIPDEW
MMAR_4003|M.marinum_M LVHTNNWDPAVLQRFREDPVVTSVRG--GIDHKATAEQIEHIATLLPDEW
MUL_3866|M.ulcerans_Agy99 LVHTNNWDPAVLQRFREDPVVTSVRG--GIDHKATAEQIEHIATLLPDEW
MAV_5061|M.avium_104 LARVNGWSLAPVERLRAAVAKVKSAGDELVDTAFHRIQLMDLADMVPDEW
MAB_2816c|M.abscessus_ATCC_199 VARYGSQGDVP------QTLTAYIAGRDGYDYNQHGRAGNTHAEFVSDDI
MLBr_00269|M.leprae_Br4923 ------------------------AAEQKHSIDDPIEMEKVADALPIEQV
. .:
Mb3120c|M.bovis_AF2122/97 LDRVCALGSPDRVAARLRAYADAGADCVAVVPATAEDPGGRVALRALRPG
Rv3093c|M.tuberculosis_H37Rv LDRVCALGSPDRVAARLRAYADAGADCVAVVPATAEDPGGRVALRALRPG
MSMEG_2249|M.smegmatis_MC2_155 LEP-SATGSAQQCVARIRKEFDYGADAVIMHGATPDELEPIVAEYRATR-
TH_1627|M.thermoresistible__bu LEP-AATGSARQCVERIRKEFDYGADAVIMHGATPDELEPIVAEYRAVVG
Mflv_4328|M.gilvum_PYR-GCK LEP-SATGSPQQCADRVRKEFDYGADAVIMHGATPDELEPIVAAYRAGQS
Mvan_2018|M.vanbaalenii_PYR-1 LAP-SATGSPQQCADRVRQELDYGADAVIMHGATPDELEPIVAAYRAGRS
MMAR_4003|M.marinum_M LEP-SATGSAQQCVDRIRAEFGYGADAVIMHGATPDELEPVVAAYRQKAQ
MUL_3866|M.ulcerans_Agy99 LEP-SATGSAQQCVDRIRAEFGYGADAVIMHGATPDELEPVVAAYRQKAQ
MAV_5061|M.avium_104 MEASCAFGTAAQCARKLQEFKRAGADEIATYGSTPSQNAGVLAAWSDAFA
MAB_2816c|M.abscessus_ATCC_199 VDRFCVLGTADEHIAKLSELKALGVDQFAGYLQHDNKEETLRVYGETVIP
MLBr_00269|M.leprae_Br4923 AKRWIVVSDPDEAVARVGQYVTWGLNHLVFHAPGHNQRRFLELFEKDLAP
. . . . :: * : . ..
Mb3120c|M.bovis_AF2122/97 GLYGTAGDNDGRR
Rv3093c|M.tuberculosis_H37Rv GLYGTAGDNDGRR
MSMEG_2249|M.smegmatis_MC2_155 -------------
TH_1627|M.thermoresistible__bu -------------
Mflv_4328|M.gilvum_PYR-GCK -------------
Mvan_2018|M.vanbaalenii_PYR-1 -------------
MMAR_4003|M.marinum_M -------------
MUL_3866|M.ulcerans_Agy99 -------------
MAV_5061|M.avium_104 GDMR---------
MAB_2816c|M.abscessus_ATCC_199 ALSQHITAKSG--
MLBr_00269|M.leprae_Br4923 RLRRLG-------