For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDDGQAGGVALGAYVLPGRVTDPGAVVSQAEAAERLGLRTVWLSERWGTKDLGVIAGALSQVTTSIRIAS GITHLQSRHPALLASLAMTAQALSGGRFILGVGRSVDAMWTAVGLPRSTNASIVDHADIFRRLCSGEKVR YEGPAGTYPSLRLIDVPNQPVPPLVFAAIGPKGLELAGRHFDGVLLHPFLTPAAVRRSVDLVRAAERTAG RPKGSVRVYATVVVACELLAEEELAVVGARAVTYYQIPGFGERLAAVNGWDPEPLARLRSHPLLAGIRGA ADSVLTREDLVEVASVLPAAWTGDAAAIGSARACHSMFERYREAGADELVLHGSTPDRLGPLLPTTKAGS AHE
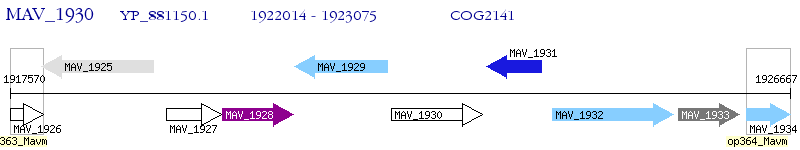
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1930 | - | - | 100% (353) | hypothetical protein MAV_1930 |
| M. avium 104 | MAV_1607 | - | 3e-60 | 38.60% (342) | hypothetical protein MAV_1607 |
| M. avium 104 | MAV_5061 | - | 7e-58 | 38.17% (317) | hypothetical protein MAV_5061 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0137c | fgd2 | 1e-13 | 30.41% (171) | putative f420-dependant glucose-6-phosphate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4328 | - | 6e-59 | 36.92% (344) | luciferase family protein |
| M. tuberculosis H37Rv | Rv0132c | fgd2 | 1e-13 | 30.41% (171) | putative f420-dependent glucose-6-phosphate dehydrogenase |
| M. leprae Br4923 | MLBr_00348 | - | 9e-11 | 28.06% (196) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_2053 | - | 4e-16 | 26.27% (335) | luciferase-like |
| M. marinum M | MMAR_4003 | - | 8e-62 | 39.29% (336) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2249 | - | 3e-61 | 38.42% (341) | hypothetical protein MSMEG_2249 |
| M. thermoresistible (build 8) | TH_1627 | - | 1e-60 | 38.71% (341) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3866 | - | 3e-62 | 39.58% (336) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_2018 | - | 2e-58 | 36.84% (342) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4328|M.gilvum_PYR-GCK -----------------------------MTEAKMDELGYYLLAGAGGEG
Mvan_2018|M.vanbaalenii_PYR-1 -----------------------------MTR-VLDELGFYLLAGAGGEG
MSMEG_2249|M.smegmatis_MC2_155 MNSRLFNSHRVVAGCVTECGIVVTNEEFDMSEHALDELGYYLLAGAGGEG
TH_1627|M.thermoresistible__bu -----------------------------MNR-PLDELGYYLLAGAGGEG
MMAR_4003|M.marinum_M -----------------------------MSDRTLNELGYYLLAGAGGEG
MUL_3866|M.ulcerans_Agy99 -----------------------------MSDRTLNELGYYLLAGAGGEG
MAV_1930|M.avium_104 ---------------------------MDDGQAGGVALGAYVLPGR-VTD
Mb0137c|M.bovis_AF2122/97 --------------------------MTGISRRTFGLAAGFGAIGAGGLG
Rv0132c|M.tuberculosis_H37Rv --------------------------MTGISRRTFGLAAGFGAIGAGGLG
MAB_2053|M.abscessus_ATCC_1997 -----------------------------MSTYGVSVLG---------AD
MLBr_00348|M.leprae_Br4923 -----------------------------MVERANAMKLGLQLGYWAAQP
Mflv_4328|M.gilvum_PYR-GCK PATLMDEARR----GEELGFGTGFISERWNVKEASSLTGAALAVTSRMQI
Mvan_2018|M.vanbaalenii_PYR-1 PATLMDEARR----GEELGFGTGFISERWNVKEASSLTGAALAVTSRMQI
MSMEG_2249|M.smegmatis_MC2_155 PATLMDEARR----GEELGFGTAFISERWNVKEASSLVGAACAVTTRMQI
TH_1627|M.thermoresistible__bu PATLMDEARR----GEELGFGTAFISERWNVKEASSLVGAACAVTERMQI
MMAR_4003|M.marinum_M PATLMDEARR----GEELGLGTAFVSERWNVKEASSLVGAACAVTSRMQI
MUL_3866|M.ulcerans_Agy99 PATLMDEARR----GEELGLGTAFVSERWNVKEASSLVGAACAVTSRMQI
MAV_1930|M.avium_104 PGAVVSQAEA----AERLGLRTVWLSERWGTKDLGVIAGALSQVTTSIRI
Mb0137c|M.bovis_AF2122/97 GGCSTRSGPTPTPEPASRGVGVVLSHEQFRTDRLVAHAQAAEQAGFRYVW
Rv0132c|M.tuberculosis_H37Rv GGCSTRSGPTPTPEPASRGVGVVLSHEQFRTDRLVAHAQAAEQAGFRYVW
MAB_2053|M.abscessus_ATCC_1997 LATLVETAEA----TDLAGFDAAWASEFYSRSGSISMA-AMAARTKRCRI
MLBr_00348|M.leprae_Br4923 PGNAAELVAA----AEGAGFDAVFTGEAWG-SDAYTPLAWWGSSTQRVRL
. *. . * : .
Mflv_4328|M.gilvum_PYR-GCK ATAATN----HNTRHPLITGSWATTMHRLSGGRFTLGIG---RGIGAIYN
Mvan_2018|M.vanbaalenii_PYR-1 ATAATN----HNTRHPLITGSWATTMHRLSGGRFTLGIG---RGIGAIYN
MSMEG_2249|M.smegmatis_MC2_155 ATAATN----HNTRHPLITGSWATTMHRLSGGRFTLGIG---RGIAAMYG
TH_1627|M.thermoresistible__bu ATAATN----HNTRHPLITGSWATTMHRLSGGRFTLGIG---RGVAAMYA
MMAR_4003|M.marinum_M ATAATN----HNTRHPLVTASWATTMHRLSGGRFTLGIG---RGIAAIYG
MUL_3866|M.ulcerans_Agy99 ATAATN----HNTRHPVVTASWATTMHRLSGGRFTLGIG---RGIAAIYG
MAV_1930|M.avium_104 ASGITH----LQSRHPALLASLAMTAQALSGGRFILGVG---RSVDAMWT
Mb0137c|M.bovis_AF2122/97 ASDHLQPWQDNEGHSMFPWLTLALVGNSTSSILFGTGVT---CPIYRYHP
Rv0132c|M.tuberculosis_H37Rv ASDHLQPWQDNEGHSMFPWLTLALVGNSTSSILFGTGVT---CPIYRYHP
MAB_2053|M.abscessus_ATCC_1997 GSSILYG----VGRSPLVLATEARDLDELSGGRVVLGLGNGTRRMMSDWH
MLBr_00348|M.leprae_Br4923 GTSVVQ----LSARTPTACAMAALTLDHLSGGRHILGLGVSGPQVVEGWY
.: : * . *. *: :
Mflv_4328|M.gilvum_PYR-GCK AFGIPTVTTAQMEDFAQVMRKLWH-GELIFNHDGPLGKYQVLFLDPDFR-
Mvan_2018|M.vanbaalenii_PYR-1 AFGIPTVTTAQMEDFAQVMRKLWH-GELIFNHDGPLGKYQVLFLDPDFR-
MSMEG_2249|M.smegmatis_MC2_155 AFGIPSVTTAQMEDFAQVMRRLWQ-GEVIFNHDGPIGNYQVLFLDPDFR-
TH_1627|M.thermoresistible__bu AFGIPPVTTAQMEDFAQVMRRLWR-GEVIFDHDGPIGRYQVLFLDPDFN-
MMAR_4003|M.marinum_M AFGIPAVTTAQMEDWAQVMRRLWK-GEMIFNHDGPMGRYPVLFLDPDFN-
MUL_3866|M.ulcerans_Agy99 AFGIPAVTTAQMEDWAQVMRRLWK-GEMVFNHDGPMGRYPVLFLDPDFN-
MAV_1930|M.avium_104 AVGLPRSTNASIVDHADIFRRLCS-GEKVR-YEGPAGTYPSLRLIDVPN-
Mb0137c|M.bovis_AF2122/97 ATVAQAFASLAILNPGRVFLGLGT-GKRLN-EQAATDTFGNYRERHDRL-
Rv0132c|M.tuberculosis_H37Rv ATVAQAFASLAILNPGRVFLGLGT-GERLN-EQAATDTFGNYRERHDRL-
MAB_2053|M.abscessus_ATCC_1997 GVEDTSAPALRMEELVPLVRRIWNLHEGPVRHEGRFYRMNLVPTGDVAPP
MLBr_00348|M.leprae_Br4923 GQPFP-KPLSRTREYIDIVRQVWA-REAPVVSDGQHYPLPLTGAGATGLG
. . : :. : : :.
Mflv_4328|M.gilvum_PYR-GCK EDIR-LAIVAFGPQTLALGGREFDDVILHTYFTPETLQRAVK-TVKDAAE
Mvan_2018|M.vanbaalenii_PYR-1 EDIR-LAIVAFGPQTLALGGREFDDVILHTYFTPETLQRAVR-TVKDAAE
MSMEG_2249|M.smegmatis_MC2_155 EDIR-LAIVAFGPQTLALGGREFDDVILHTYFTPETLQRAVK-TVKTAAE
TH_1627|M.thermoresistible__bu EDIR-LAIVAFGPQTLALGGRMFDDVILHTYFTPETLQRAVK-TVKDAAE
MMAR_4003|M.marinum_M EDIR-LALVALGPKSLALGGRVFDDVILHTYFTPETLQRCVS-IVKTAAD
MUL_3866|M.ulcerans_Agy99 EDIR-LALVALGPKSLALGGRVFDDVILHTYFTPETLQRCVS-IVKTAAD
MAV_1930|M.avium_104 QPVPPLVFAAIGPKGLELAGRHFDGVLLHPFLTPAAVRRSVD-LVRAAER
Mb0137c|M.bovis_AF2122/97 IEAIVLIRQLWSGERISFTGHYFRTDELKLYDTPAMPPPIFV-AASGPQS
Rv0132c|M.tuberculosis_H37Rv IEAIVLIRQLWSGERISFTGHYFRTDELKLYDTPAMPPPIFV-AASGPQS
MAB_2053|M.abscessus_ATCC_1997 QRAIPIITAGVRPRMCEVAGRVADGLAGHPLFTTTYVEEVARPAVVRGAE
MLBr_00348|M.leprae_Br4923 KALKPITHPLRADIPIMLGAEGPKNVALAAEICDGWLPIFFSPRMADMYN
: . ..
Mflv_4328|M.gilvum_PYR-GCK QAGRDPASVRVWSCFATVGDHLP-EELRLKKTVARLATYLQGYGDLLVNT
Mvan_2018|M.vanbaalenii_PYR-1 QAGRDPASVRVWSCFATVGDHLP-EELRLKKTVARLATYLQGYGDLLVNT
MSMEG_2249|M.smegmatis_MC2_155 QAGRDPGSVRVWSCFATVGDHLP-EELRLKKTVARLATYLQGYGDLLVNT
TH_1627|M.thermoresistible__bu QAGRDPAEVRVWSCFATVGDHLP-EELRLKKTVARLATYLQGYGDLLVET
MMAR_4003|M.marinum_M RAGRDPASVRVWSCFATVGDHLP-EQLRLKKTVARLATYLQGYGDLLVHT
MUL_3866|M.ulcerans_Agy99 RAGRDPASVRVWSCFATVGDHLP-EQLRLKKTVARLATYLQGYGDLLVHT
MAV_1930|M.avium_104 TAGRPKGSVRVYATVVVACELLAEEELAVVGARAVTYYQIPGFGERLAAV
Mb0137c|M.bovis_AF2122/97 ATLAGRYGDGWIAQARDINDAKLLAAFAAGAQAAGRDPTTLGKRAELFAV
Rv0132c|M.tuberculosis_H37Rv ATLAGRYGDGWIAQARDINDAKLLAAFAAGAQAAGRDPTTLGKRAELFAV
MAB_2053|M.abscessus_ATCC_1997 RAGRDPADIEIVSMVICAVHDDPQIARREAAQQIAFYSSVKTYEHVLDVS
MLBr_00348|M.leprae_Br4923 EWLDEGFARTGARRSREDFEICASAQIVVTEDRAAAFAGIKPFLALYMGG
.
Mflv_4328|M.gilvum_PYR-GCK --NGWDPAVLQRFRDDKVVQSIGGGIDHKATAEQIEHIATLIPDEWL-EP
Mvan_2018|M.vanbaalenii_PYR-1 --NGWDPAVLQRFRDDKVVQSVGGGIDHKATAEQIEHIATLIPDEWL-AP
MSMEG_2249|M.smegmatis_MC2_155 --NGWDPAVLQRFRDDKVVQSIPGGIDHKATAEQIEHIATLIPDEWL-EP
TH_1627|M.thermoresistible__bu --NGWDPAVLRKFRDDPVVQSIPGGIDHKATPEQIEHIATLIPDEWL-EP
MMAR_4003|M.marinum_M --NNWDPAVLQRFREDPVVTSVRGGIDHKATAEQIEHIATLLPDEWL-EP
MUL_3866|M.ulcerans_Agy99 --NNWDPAVLQRFREDPVVTSVRGGIDHKATAEQIEHIATLLPDEWL-EP
MAV_1930|M.avium_104 --NGWDPEPLARLRSHPLLAGIRGAADSVLTREDLVEVASVLPAAWTGDA
Mb0137c|M.bovis_AF2122/97 --VGDDK---AAARAADLWRFTAGAVDQ-PNPVEIQRAAESNPIEKVLAN
Rv0132c|M.tuberculosis_H37Rv --VGDDK---AAARAADLWRFTAGAVDQ-PNPVEIQRAAESNPIEKVLAN
MAB_2053|M.abscessus_ATCC_1997 GFARQGAAIRDAFARRDLPAMFAAVTDDMIDAMAVAGTAAEVREGLR---
MLBr_00348|M.leprae_Br4923 -MGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAEIIPNELVDST
. *
Mflv_4328|M.gilvum_PYR-GCK SATGSPQQCADRVRKEFDYGADAVIMHGATPDELEPIVAAYRAGQS---
Mvan_2018|M.vanbaalenii_PYR-1 SATGSPQQCADRVRQELDYGADAVIMHGATPDELEPIVAAYRAGRS---
MSMEG_2249|M.smegmatis_MC2_155 SATGSAQQCVARIRKEFDYGADAVIMHGATPDELEPIVAEYRATR----
TH_1627|M.thermoresistible__bu AATGSARQCVERIRKEFDYGADAVIMHGATPDELEPIVAEYRAVVG---
MMAR_4003|M.marinum_M SATGSAQQCVDRIRAEFGYGADAVIMHGATPDELEPVVAAYRQKAQ---
MUL_3866|M.ulcerans_Agy99 SATGSAQQCVDRIRAEFGYGADAVIMHGATPDELEPVVAAYRQKAQ---
MAV_1930|M.avium_104 AAIGSARACHSMFERYREAGADELVLHGSTPDRLGPLLPTTKAGSAHE-
Mb0137c|M.bovis_AF2122/97 WAVGTDPGVHIGAVQAVLDAGAVPFLHFPQDDPITAIDFYRTNVLPELR
Rv0132c|M.tuberculosis_H37Rv WAVGTDPGVHIGAVQAVLDAGAVPFLHFPQDDPITAIDFYRTNVLPELR
MAB_2053|M.abscessus_ATCC_1997 RYEGVLDHIVLYSPSIGLAPERIAENLGSLIRDCAPAFAGGKGDQSG--
MLBr_00348|M.leprae_Br4923 MIVGDVDYVCKQIAAWSAAGVTMMMVSASSVEQVRDLAGLF--------
* .