For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTAQLASHLTRALTLAQQQPYLARRQNWVNQLERHAMMQPDAPALRFVGNTMTWADLRRRVAALAGALSG RGVGFGDRVMILMLNRTEFVESVLAANMIGAIAVPLNFRLTPTEIAVLVEDCAAHVMLTEAALAPVAIGV RNIQPLLSVIVVAGGSSQDSVFGYEDLLNEAGDVHEPVDIPNDSPALIMYTAGTTGRPKGAVLTHANLTG QAMTALYTSGANINSDVGFVGVPLFHIAGIGNMLTGLLLGLPTVIYPLGAFDPGQLLDVLEAEKVTGIFL VPAQWQAVCTEQQARPRDLRLRVLSWGAAPAPDALLRQMSATFPETQILAAFGQTEMSPVTCMLLGEDAI AKRGSVGRVIPTVAARVVDQNMNDVPVGEVGEIVYRAPTLMSCYWNNPEATAEAFAGGWFHSGDLVRMDS DGYVWVVDRKKDMIISGGENIYCAELENVLASHPDIAEVAVIGRADEKWGEVPIAVAAVTNDDLRIEDLG EFLTDRLARYKHPKALEIVDALPRNPAGKVLKTELRLRYGACVNVERRSASAGFTERRENRQKL
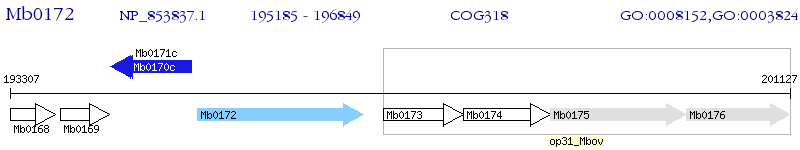
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0172 | fadD5 | - | 100% (554) | acyl-CoA synthetase |
| M. bovis AF2122 / 97 | Mb3116 | fadD13 | 3e-73 | 36.14% (487) | chain-fatty-acid-CoA ligase |
| M. bovis AF2122 / 97 | Mb2533c | fadD35 | 1e-59 | 34.36% (486) | AMP-binding domain protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0714 | - | 0.0 | 70.95% (537) | acyl-CoA synthetase |
| M. tuberculosis H37Rv | Rv0166 | fadD5 | 0.0 | 99.64% (554) | acyl-CoA synthetase |
| M. leprae Br4923 | MLBr_02661 | - | 1e-39 | 29.38% (514) | acyl-CoA synthetase |
| M. abscessus ATCC 19977 | MAB_4569c | - | 0.0 | 73.24% (512) | fatty-acid-CoA ligase FadD |
| M. marinum M | MMAR_0409 | fadD5 | 0.0 | 82.45% (547) | fatty-acid-CoA ligase FadD5 |
| M. avium 104 | MAV_5018 | - | 0.0 | 82.83% (530) | acyl-CoA synthetase |
| M. smegmatis MC2 155 | MSMEG_0131 | - | 0.0 | 72.37% (532) | acyl-CoA synthetase |
| M. thermoresistible (build 8) | TH_2878 | - | 0.0 | 73.73% (533) | PUTATIVE AMP-binding enzyme, putative |
| M. ulcerans Agy99 | MUL_0650 | fadD8 | 2e-54 | 32.05% (521) | acyl-CoA synthetase |
| M. vanbaalenii PYR-1 | Mvan_0131 | - | 0.0 | 74.38% (523) | acyl-CoA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0714|M.gilvum_PYR-GCK MTRTTSCLIKCHTSANPSTMERKTLTDPTGLSGSATEQPNLARRRNWANQ
Mvan_0131|M.vanbaalenii_PYR-1 -------MFDQISSFRNPSMEQKTLTEAIQQPGFATEQPYLARRQNWTNQ
MSMEG_0131|M.smegmatis_MC2_155 ----------------------------MTAEPSQIEQPYLARRQNWVNQ
TH_2878|M.thermoresistible__bu -----------------------------------LQQPYLARRQNWVNH
Mb0172|M.bovis_AF2122/97 -------------------MTAQLASHLTRALTLAQQQPYLARRQNWVNQ
Rv0166|M.tuberculosis_H37Rv -------------------MTAQLASHLTRALTLAQQQPYLARRQNWVNQ
MMAR_0409|M.marinum_M -------------------MTAELASHLPQAA-HALEQPYLARRQNWVNQ
MAV_5018|M.avium_104 --------------------MLSLTAQLAHHPTQANEQPYLSRRQNWVNQ
MAB_4569c|M.abscessus_ATCC_199 -------------------MTDTIDATVQSAG---LAQPYRARRQNWCNQ
MUL_0650|M.ulcerans_Agy99 ------------------------------------MSDDLLRNPTHNGH
MLBr_02661|M.leprae_Br4923 ------------------MKSESITSSRAPVEQIDARLTGQFESPSIASL
. . .
Mflv_0714|M.gilvum_PYR-GCK LSRHALMQPDTVALRFLG--RSTTWGELDRRVTALAGALHERGVGFGDRV
Mvan_0131|M.vanbaalenii_PYR-1 LARHALMQPDQPALRFLG--NTTTWGELDRRVSALAGALHKRGVGFGDRV
MSMEG_0131|M.smegmatis_MC2_155 LTRHAMMQPDKTALRFLG--RTTTWRELDDRVTRLAGALNRRGVGFGDRV
TH_2878|M.thermoresistible__bu LARHALMQPDATALRFMG--QTTTWADLHHRVNRLAGALRRRGVEFGDRV
Mb0172|M.bovis_AF2122/97 LERHAMMQPDAPALRFVG--NTMTWADLRRRVAALAGALSGRGVGFGDRV
Rv0166|M.tuberculosis_H37Rv LERHAMMQPDAPALRFVG--NTMTWADLRRRVAALAGALSGRGVGFGDRV
MMAR_0409|M.marinum_M LERHAMMQPRATAIRYLG--HTVTWADLRYRVAALAGSLSRRGVGFGDRV
MAV_5018|M.avium_104 LERHALMQPNATALRFLG--KGLTWGELHGRVRALADALSRRGVGFGDRV
MAB_4569c|M.abscessus_ATCC_199 LARHALMQPDATAIRYLG--HSISWAELHQRVQALADALSRRGVGFGDRV
MUL_0650|M.ulcerans_Agy99 LLVGALERHKNRPVLFLGD-TTLTGGQLAERISQYIQAFEALGAGTGVTV
MLBr_02661|M.leprae_Br4923 VEAAAIRNPSAPALVVTDNRIVVSYRDLLRLVDDLTVQLALGGLLPGDRV
: *: . .: . : :* : : * * *
Mflv_0714|M.gilvum_PYR-GCK LILMLNRTEFMESVLAVNKLGGIAVPVNFRMTPSEIAFLVSDCAAEVVIT
Mvan_0131|M.vanbaalenii_PYR-1 LILMLNRTEFIEAVLATNKLGAIAVPVNFRMTPAEIAFLVSDCEAKAIIT
MSMEG_0131|M.smegmatis_MC2_155 LILMLNRTEFVEAMLAANRLGAIAVPVNFRMTPPEIAYLVSDCEAHVVVT
TH_2878|M.thermoresistible__bu LILMLNRTEFIETMLAANRLGAIAVPVNFRMTPPEIAFLAKDCEAKVIAT
Mb0172|M.bovis_AF2122/97 MILMLNRTEFVESVLAANMIGAIAVPLNFRLTPTEIAVLVEDCAAHVMLT
Rv0166|M.tuberculosis_H37Rv MILMLNRTEFVESVLAANMIGAIAVPLNFRLTPTEIAVLVEDCVAHVMLT
MMAR_0409|M.marinum_M MILMLNRTEFVESVLAANMLGAIAVPLNFRLTPSEIAFLVEDCAPRLVIT
MAV_5018|M.avium_104 MVLMLNRPEFMESVLAINMLGAIAVPLNFRLTAAEIAFLVQDCQARVVIT
MAB_4569c|M.abscessus_ATCC_199 LILMLNRPEYVESWLAINELGAIAIPVNFRMTPPEVAFLVENSGAKVAIT
MUL_0650|M.ulcerans_Agy99 GLLSLNRPEVLMILGASQARGYRRTALHPLGSLDDHAYVLYDAGASALII
MLBr_02661|M.leprae_Br4923 ALCAASNIEFVVGLLAASRAGLIVVPLDPALPVNEQCIRSQAAGVRVTLV
: .. * : * . * .:. . : . .
Mflv_0714|M.gilvum_PYR-GCK ES--VLENVATAVRDIDPKLATVIVAG---GTSRDNVLDYDDVLASTT-A
Mvan_0131|M.vanbaalenii_PYR-1 EA--VLANVATAVRDIDPTLATVISAG---GSG-DDVLDYDDVLAENA-P
MSMEG_0131|M.smegmatis_MC2_155 EP--VLAGVATAVRDLDATLQTVIVAGGATEDG---VLGYDDLIAEEG-E
TH_2878|M.thermoresistible__bu EP--VLAGVATAVRDLAPSLSSVIVAGAADGSDTPDVLDYEALIAEPG-E
Mb0172|M.bovis_AF2122/97 EA--ALAPVAIGVRNIQPLLSVIVVAG---GSSQDSVFGYEDLLNEAG-D
Rv0166|M.tuberculosis_H37Rv EA--ALAPVAIGVRNIQPLLSVIVVAG---GSSQDSVFGYEDLLNEAG-D
MMAR_0409|M.marinum_M EE--VLAQVAVGVREIAPALSTIVVAG---GASDPTVVGYDELISETG-D
MAV_5018|M.avium_104 EA--VLAPVATGVRDIESLLDTVVVAG---GSSDDAVLGYDDLIDETG-A
MAB_4569c|M.abscessus_ATCC_199 ET--VLAPVAAAVRQQAPALETVIVAG---SASENGVLGYEEVIAEPG-E
MUL_0650|M.ulcerans_Agy99 DPNPMFVERALGLLEKVDSLEQVLTIGPVPEALEDRAVDLSAEAAKYQPK
MLBr_02661|M.leprae_Br4923 DS-LALEGVSDQRAATMRYWPIAVSYGSVTGASEGSLLVHLDDTAALHPV
: : : : * .
Mflv_0714|M.gilvum_PYR-GCK APPLVDIPDDSPALIMYTSGTTGRPKGAVLTHNNLAGQAMTLLFTNGADI
Mvan_0131|M.vanbaalenii_PYR-1 CP-VVDIPNDAPALIMYTSGTTGRPKGAVLTHTNLTGQVMTLLFTNGADI
MSMEG_0131|M.smegmatis_MC2_155 PPAPVDIPNDSPALIMYTSGTTGRPKGAVLTHTNLSAQAMMHLFTTGADL
TH_2878|M.thermoresistible__bu DPPPVDIPDDSPALIMYTSGTTGNPKGAVLTHSNLTGQAMTNLFTIGVDL
Mb0172|M.bovis_AF2122/97 VHEPVDIPNDSPALIMYTAGTTGRPKGAVLTHANLTGQAMTALYTSGANI
Rv0166|M.tuberculosis_H37Rv VHEPVDIPNDSPALIMYTSGTTGRPKGAVLTHANLTGQAMTALYTSGANI
MMAR_0409|M.marinum_M PPEPVDIPNDSPALIMYTSGTTGRPKGAVLTHTNLTGQVMTALYTGGANI
MAV_5018|M.avium_104 AHQPVDIPNDAAALIMYTSGTTGRPKGAVLTHTNLTGQTMTGLYTNGADI
MAB_4569c|M.abscessus_ATCC_199 PHAEVDLPGDTPALIMYTSGTTGRPKGAVLTHLNLSGQAMTYLLSTTVDL
MUL_0650|M.ulcerans_Agy99 PLVVADLPPNHIGGLTYTGGTTGKPKGVIGTAGNIT--AMTSIQLAEWEW
MLBr_02661|M.leprae_Br4923 TSTPDGLRHDD-AMIMFTGGTTGLPKMVPWTDGNIAGSVHAIITAYQLGP
.: : . : :*.**** ** . * *:: . :
Mflv_0714|M.gilvum_PYR-GCK NHDIGFIGVPFFHIAGVG-SIVSGMMLGRPTVIYPLGAFDPNELLDVLEA
Mvan_0131|M.vanbaalenii_PYR-1 NHDIGFIGVPFFHIAGVG-SSVSGLLLGRPTVLYPVGAFDPNELLDVLEA
MSMEG_0131|M.smegmatis_MC2_155 NSDVGFVGVPLFHIAGVAGSVLSGVTLGVPTVLYPLGAFDPDGLLDVLES
TH_2878|M.thermoresistible__bu ANDVGFIGVPLFHIAGIAGNVLSGLTLGLPTVLYPLGAFDPGGLLDVLEA
Mb0172|M.bovis_AF2122/97 NSDVGFVGVPLFHIAGIG-NMLTGLLLGLPTVIYPLGAFDPGQLLDVLEA
Rv0166|M.tuberculosis_H37Rv NSDVGFVGVPLFHIAGIG-NMLTGLLLGLPTVIYPLGAFDPGQLLDVLEA
MMAR_0409|M.marinum_M NSDVGFIGVPFFHIAGIG-NMLSGMLLGVPTVIYPLGAFNPGQLLDVLEA
MAV_5018|M.avium_104 NNDVGFIGVPFFHIAGIG-NMLTGLLLGIPTVIYPLGAFEPGQLLDVLAA
MAB_4569c|M.abscessus_ATCC_199 NSDVGFIGVPMFHIAGVG-NMITGLLLGLPTVIHPLGAFDPGALLDVLES
MUL_0650|M.ulcerans_Agy99 PEHPRFLMCTPLSHAGAAFFTPTLVKGGE---MIVLAKFDPGEVLRVIEE
MLBr_02661|M.leprae_Br4923 -QDATVVVMPLYHGHGLIAALLSTLASGGVVLLPARARFSARTFWDDIDA
. .: . * : : * : . *.. . :
Mflv_0714|M.gilvum_PYR-GCK EKVTGIFLVPAQWQAVCAAQKANPRNLK---LRFLSWGAAPASDTLLRQM
Mvan_0131|M.vanbaalenii_PYR-1 EKVTGIFLVPAQWQAVCAAQKARPRDLK---LRFLSWGAAPASDTLLREM
MSMEG_0131|M.smegmatis_MC2_155 EQVTGIFLVPAQWQAVCAAQRARPRNLK---LRALSWGAAPASDTLLREM
TH_2878|M.thermoresistible__bu EKVTGIFLVPAQWQAVCAEQRARPRDLR---LRALSWGAAPASDTLLQEM
Mb0172|M.bovis_AF2122/97 EKVTGIFLVPAQWQAVCTEQQARPRDLR---LRVLSWGAAPAPDALLRQM
Rv0166|M.tuberculosis_H37Rv EKVTGIFLVPAQWQAVCTEQQARPRDLR---LRVLSWGAAPAPDALLRQM
MMAR_0409|M.marinum_M ERVSGIFLVPAQWQAVCAEQQARPRELS---LRVMSWGAAPAPDALLRQM
MAV_5018|M.avium_104 EKVTGIFLVPAQWQAVCAEQRARPRDLK---LRVISWGAAPAPDALLREM
MAB_4569c|M.abscessus_ATCC_199 EGVTGIFLVPAQWQAVCAAQQANPRKVR---LKTLSWGAAPATDVLLRTM
MUL_0650|M.ulcerans_Agy99 QRITATMLVPSMLYALMDHPDPHTRDLSS--LETVYYGASAINPVRLAEA
MLBr_02661|M.leprae_Br4923 VAATWYTAAPAIHRILLELASTQSFRSKRAKLRFIRSCSAPLTQETAQAL
: .*: : ... *. : ::.
Mflv_0714|M.gilvum_PYR-GCK SETFPGAQILAA-----FGQTEMSPVTCMLLGDDALRKLGSVGKVIPTVS
Mvan_0131|M.vanbaalenii_PYR-1 AETFPGAQILAA-----FGQTEMSPVTCMLLGDDAIRKLGSVGKVIPTVS
MSMEG_0131|M.smegmatis_MC2_155 NEVFPDAQIFAA-----FGQTEMSPVTCMLLGDDAIRKLGSVGRVIPTVS
TH_2878|M.thermoresistible__bu TEVFPGTKIFAA-----FGQTEMSPVTCMLLAEDAVRKLGSVGKVIPTVA
Mb0172|M.bovis_AF2122/97 SATFPETQILAA-----FGQTEMSPVTCMLLGEDAIAKRGSVGRVIPTVA
Rv0166|M.tuberculosis_H37Rv SATFPETQILAA-----FGQTEMSPVTCMLLGEDAIAKRGSVGRVIPTVA
MMAR_0409|M.marinum_M SEVFPGTQIMAA-----FGQTEMSPVTCMLLGEDAIRKRGSVGKVIPTVS
MAV_5018|M.avium_104 SAMFPGTQILAA-----FGQTEMSPVTCMLLGEDAIRKRGSVGKVIPTVA
MAB_4569c|M.abscessus_ATCC_199 SETFPDARILAA-----FGQTEMSPVTCMLMGDDAIRKMGSVGRVIPTVS
MUL_0650|M.ulcerans_Agy99 IDRFGPIFAQYY-----GQSEAPMVITYLPKADHDEKRLTSCGRPTLFAR
MLBr_02661|M.leprae_Br4923 REEFLAPVICAFGMTEATHQVTTTNIKWFGQGENPTVTNGLVGQSTG-VQ
* . :. : .:. *: .
Mflv_0714|M.gilvum_PYR-GCK ARIVDEDMNDVPIGQVGEIVYRAPTLMTGYWNNPTATAEAFAGGWFHSGD
Mvan_0131|M.vanbaalenii_PYR-1 ARIVDEDMNDVPVGQVGEIVYRAPTLMAGYWNNPKATAEAFAGGWFHSGD
MSMEG_0131|M.smegmatis_MC2_155 ARVVDDNMNDVPVGEVGEIVYRGPNLMVGYWNNPKATAEAFAGGWFHSGD
TH_2878|M.thermoresistible__bu ARIVDDTMNDVPVGEVGEIVYRAPTLMAGYWNNPQATAEAFAGGWFHSGD
Mb0172|M.bovis_AF2122/97 ARVVDQNMNDVPVGEVGEIVYRAPTLMSCYWNNPEATAEAFAGGWFHSGD
Rv0166|M.tuberculosis_H37Rv ARVVDQNMNDVPVGEVGEIVYRAPTLMSCYWNNPEATAEAFAGGWFHSGD
MMAR_0409|M.marinum_M ARVVDDEMNDVPIGQVGEIVYRAPTLMSGYWNNPDATAEAFAGGWFHSGD
MAV_5018|M.avium_104 ARVVDENMNDVPVGEVGEIVYRAPTLMSGYWNNPEATAEAFAGGWFHSGD
MAB_4569c|M.abscessus_ATCC_199 ARVVDDNMNDVPVGEVGEIVYRAPTLMQGYWNNPEATADAFAGGWFHSGD
MUL_0650|M.ulcerans_Agy99 VALLGEDGKPVPQGEPGEICVSGPLLAGGYWNKPDDTAQTFRDGWLHTGD
MLBr_02661|M.leprae_Br4923 IRIVGSDGQPLPPDTVGEVWLRGSTVVRGYLGDPAITAANFTHGWLRTGD
::.. : :* . **: .. : * ..* ** * **:::**
Mflv_0714|M.gilvum_PYR-GCK LVRQDEEGYIWVVDRKKDMIISGGENIYCAEVENVLASHPDIVEVAVIGR
Mvan_0131|M.vanbaalenii_PYR-1 LVRQDEEGYIWVVDRKKDMIISGGENIYCAEVENVLAAHPEIVEVAVIGR
MSMEG_0131|M.smegmatis_MC2_155 LVRQDEEGYVWVVDRKKDMIISGGENIYCAEVENVLAAHPAIMEVAVIGR
TH_2878|M.thermoresistible__bu LVRQDEEGYIWVVDRKKDMIISGGENIYCAEVENVLAAHPAVAEVAVVGR
Mb0172|M.bovis_AF2122/97 LVRMDSDGYVWVVDRKKDMIISGGENIYCAELENVLASHPDIAEVAVIGR
Rv0166|M.tuberculosis_H37Rv LVRMDSDGYVWVVDRKKDMIISGGENIYCAELENVLASHPDIAEVAVIGR
MMAR_0409|M.marinum_M LVRMDEDGYVWVVDRKKDMIISGGENVYCAEVENVLASHPSIVEVAVIGR
MAV_5018|M.avium_104 LVRMDEDGYVWVVDRKKDMIISGGENIYCAEVENVLASHPDIVEVAVIGR
MAB_4569c|M.abscessus_ATCC_199 LVRQDEDGFVWVVDRKKDMIISGGENIYCAEVENALAEHEQIAEVAVIGR
MUL_0650|M.ulcerans_Agy99 MAREDEDGFYFIVDRVKDMIVTGGFNVFPREVEDVVAEHSAVAQVCVVGA
MLBr_02661|M.leprae_Br4923 LGSLSVTGDLRIRGRIKELINRSGEKISPERVEGVLASHHNVMEVAVFGD
: . * : .* *::* .* :: .:*..:* * : :*.*.*
Mflv_0714|M.gilvum_PYR-GCK SHRKWGEVPVAVAVLRSSLN-STGSGRLELTDLEQFLGDRLARYKHPKAI
Mvan_0131|M.vanbaalenii_PYR-1 AHEKWGEVPVAVAVVRSSLE-ATPGAGIELADLEQFLTERLARYKHPKAL
MSMEG_0131|M.smegmatis_MC2_155 PHEKWGEVPVAVVALNS-----ATAESISLGELDSFLTERLARYKHPKAL
TH_2878|M.thermoresistible__bu PHEKWGEVPVAVVALN-------PDAELTMAELEEFLADRLARYKHPKVL
Mb0172|M.bovis_AF2122/97 ADEKWGEVPIAVAAVTN--------DDLRIEDLGEFLTDRLARYKHPKAL
Rv0166|M.tuberculosis_H37Rv ADEKWGEVPIAVAAVTN--------DDLRIEDLGEFLTDRLARYKHPKAL
MMAR_0409|M.marinum_M ADEKWGEVPIAVAAVTK--------DHLRIEDLDEYLTERLARYKHPKAL
MAV_5018|M.avium_104 AHEKWGEVPIAVAAVAN--------DNLALEDLDEFLTERLARYKHPKAL
MAB_4569c|M.abscessus_ATCC_199 PHEKWGEVPVAVAAIHG--------AALTLAELDGFLTARLARYKHPKDL
MUL_0650|M.ulcerans_Agy99 PDEKWGEAVTAVVVLRADAARDDAAIETMTAEIQAAVKQRKGSVQSPKQV
MLBr_02661|M.leprae_Br4923 PDKVYGETVTAVIVPRE-------VIAPTPSELAVFCRDRLAAFEVPTRF
... :**. ** . :: * . : *. .
Mflv_0714|M.gilvum_PYR-GCK EIVEALPRNPAGKVLKTELRVRFGSEESVDAGESATAATSSAAAPKD-
Mvan_0131|M.vanbaalenii_PYR-1 EIVDALPRNPAGKVLKTELRIRYSAASFADAGESSTPPTISTAPQRN-
MSMEG_0131|M.smegmatis_MC2_155 EVVDALPRNPSGKVLKTELRERFGRATSLDGGESNSATTVSAG-----
TH_2878|M.thermoresistible__bu EIVDALPRNPAGKVLKTELRARFAAGPSAEKEQSSSGATTSAGSS---
Mb0172|M.bovis_AF2122/97 EIVDALPRNPAGKVLKTELRLRYGACVNVERRSASAGFTERRENRQKL
Rv0166|M.tuberculosis_H37Rv EIVDALPRNPAGKVLKTELRLRYGACVNVERRSASAGFTERRENRQKL
MMAR_0409|M.marinum_M EIVDALPRNPSGKVLKTELRLRYGVGVYSESRSVSRSFEFREGS----
MAV_5018|M.avium_104 EIVDALPRNPAGKVLKTELRIRYGGG----------------------
MAB_4569c|M.abscessus_ATCC_199 VIVDALPRNPSGKVLKTELRQRFGIAD---------------------
MUL_0650|M.ulcerans_Agy99 VVVDSLPLTGLGKPDKKAVRARFWEGAGRAVG----------------
MLBr_02661|M.leprae_Br4923 QEASALPHTAKGSLDRRAVAEQFAHRG---------------------
..:** . *. : : ::