For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSKPRALVTAPLRGPGLDKLRALAEVVYDPWIEQRPLRIYSAEQLAERIAAEEAEIVVVESDSVRGPVF ELGVRAIAATRGDPNNVDIDGATAAGIPVLNTPGRNADAVAEMTVALLLAATRHLLPADADVRGGNIFRD GSIPYQRFRGGEIAGLTAGLVGLGAVGRATRWRLSGLGLRVIAHDPYHPEARHGLDELLAESDVVSLHAP VTDETTGMIGAEQFAAMRDGVVFLNTARAQLHDTDALVEALRAGKVAAAGLDHFVGEWLPTDHPLVGMPN VVLTPHIGGATWNTEARQAQLVADDLEALLAGATPAHIVNPEVLAR
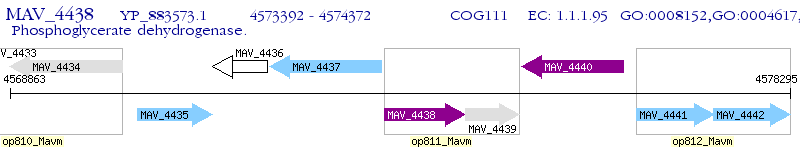
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4438 | - | - | 100% (326) | 2-hydroxyacid dehydrogenase family protein |
| M. avium 104 | MAV_3847 | serA | 1e-42 | 42.62% (244) | D-3-phosphoglycerate dehydrogenase |
| M. avium 104 | MAV_0349 | - | 3e-32 | 36.19% (257) | D-isomer specific 2-hydroxyacid dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0749c | serA2 | 1e-155 | 82.41% (324) | D-3-phosphoglycerate dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0334 | - | 1e-44 | 43.51% (239) | D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding |
| M. tuberculosis H37Rv | Rv0728c | serA2 | 1e-155 | 82.10% (324) | D-3-phosphoglycerate dehydrogenase |
| M. leprae Br4923 | MLBr_01692 | serA | 5e-43 | 42.21% (244) | D-3-phosphoglycerate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3304c | - | 2e-41 | 38.43% (281) | D-3-phosphoglycerate dehydrogenase |
| M. marinum M | MMAR_1065 | serA2 | 1e-154 | 82.98% (329) | D-3-phosphoglycerate dehydrogenase SerA2 |
| M. smegmatis MC2 155 | MSMEG_0604 | - | 1e-44 | 43.75% (240) | glyoxylate reductase |
| M. thermoresistible (build 8) | TH_3809 | - | 2e-43 | 41.94% (248) | PUTATIVE putative D-3-phosphoglycerate dehydrogenase |
| M. ulcerans Agy99 | MUL_0823 | serA2 | 1e-154 | 82.98% (329) | D-3-phosphoglycerate dehydrogenase SerA2 |
| M. vanbaalenii PYR-1 | Mvan_2128 | - | 3e-42 | 41.80% (244) | D-3-phosphoglycerate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0749c|M.bovis_AF2122/97 ---MTPRPRALVTAPLRGPGFAQLRRLADVVYDPWI---DQRPLRIYSAE
Rv0728c|M.tuberculosis_H37Rv ---MTPRPRALVTAPLRGPGFAQLRRLADVVYDPWI---DQRPLRIYSAE
MMAR_1065|M.marinum_M ---MRSRPRALVTAPLRGSGLDKLRQLVDVVYDPWIGELEGGPLRIYSAE
MUL_0823|M.ulcerans_Agy99 ---MRSRPRALVTAPLRGSGLDKLRQLVDVVYDPWIGGLEGGPLRIYSAE
MAV_4438|M.avium_104 ---MTSKPRALVTAPLRGPGLDKLRALAEVVYDPWI---EQRPLRIYSAE
TH_3809|M.thermoresistible__bu --VSGISRPRLVFERWTDPAAGELLGAADIELVRLD--LTADPADGWAAL
Mflv_0334|M.gilvum_PYR-GCK -------MRVLAHFLPGPKVTEFLSPHTDWLDIRYCA----EDDDETFYR
MSMEG_0604|M.smegmatis_MC2_155 MAVNPGTVRVLAHFAPSNTVLDFLAPHLDWLDVRFCG----EEDDETFYR
MAB_3304c|M.abscessus_ATCC_199 ------------MLIADKLAPSTVEALGDGVEVRWVD----GPDRPALLA
Mvan_2128|M.vanbaalenii_PYR-1 -------MSLPVVLIADKLAQSTVEALGDQVEVRWVD----GPDREKLLA
MLBr_01692|M.leprae_Br4923 -------MDLPVVLIADKLAQSTVAALGDQVEVRWVD----GPDRTKLLA
: :
Mb0749c|M.bovis_AF2122/97 QLADRITAVAADVLVVESD-----SVGGPVFERGLRVVAATRGD-PSNVD
Rv0728c|M.tuberculosis_H37Rv QLADRITAVAADVLVVESD-----SVGGPVFERGLRVVAATRGD-PSNVD
MMAR_1065|M.marinum_M QLAERITSESVDIVVVEAD-----SVSGAVLQLGLRAIFSTRGD-PNNVD
MUL_0823|M.ulcerans_Agy99 QLAERITSESVDIVVVEAD-----SVSGAVLQLGLRAIFSTRGD-PNNVD
MAV_4438|M.avium_104 QLAERIAAEEAEIVVVESD-----SVRGPVFELGVRAIAATRGD-PNNVD
TH_3809|M.thermoresistible__bu ESAHGYQVATRTDVASMADGGQWLADRNLVRRCGLLLAVCSAGAGYDVID
Mflv_0334|M.gilvum_PYR-GCK ELPDAEVIWH-VLRPISGD---------DLSKAPKLRLVHKLGAGVNTVD
MSMEG_0604|M.smegmatis_MC2_155 ELGDADVLWH-VLRPITGD---------DLNRAPRLRLVHKLGAGVNTID
MAB_3304c|M.abscessus_ATCC_199 AVPEADALLVRSATTVDAE---------VLAAGTKLKIVARAGVGLDNVD
Mvan_2128|M.vanbaalenii_PYR-1 AVADADALLVRSATTVDAE---------VLAAAPKLKIVARAGVGLDNVD
MLBr_01692|M.leprae_Br4923 AVPEADALLVRSATTVDAE---------VLAAAPKLKIVARAGVGLDNVD
. .: : * . :*
Mb0749c|M.bovis_AF2122/97 IPGATAAGIPVLHTPARNADAVAEMTVALLLAVARHLIPADADVRSGNIF
Rv0728c|M.tuberculosis_H37Rv IPGATAAGIPVLHTPARNADAVAEMTVALLLAVARHLIPADADVRSGNIF
MMAR_1065|M.marinum_M IAGATAAGIPVLNTPARNADAVAEMTVALLLASTRHLLTADADVRSGNIF
MUL_0823|M.ulcerans_Agy99 IAGATAAGIPVLNTPARNADAVAEMTVALLLASTRHLLTADADVRSGNIF
MAV_4438|M.avium_104 IDGATAAGIPVLNTPGRNADAVAEMTVALLLAATRHLLPADADVRGGNIF
TH_3809|M.thermoresistible__bu VDACTEAGIAVCNNSGPGAEAVAEHALGLMLDLAKRITVADRMLRRGDLG
Mflv_0334|M.gilvum_PYR-GCK VDTATARGIAVANMPGANAPSVAEGTLLLMLAALRRLPELDRATRTGNGW
MSMEG_0604|M.smegmatis_MC2_155 VETATQLGILVANMPGANAPSVAEGTVLLMLAALRRLPQLDRATRAGRGW
MAB_3304c|M.abscessus_ATCC_199 VKAATARGVLVVNAPTSNIHSAAEHAVTLLLATARQIPAADASLKAHTWK
Mvan_2128|M.vanbaalenii_PYR-1 VDAATARGVLVVNAPTSNIHSAAEHALALLLAAARQIPAADATLREHSWK
MLBr_01692|M.leprae_Br4923 VDAATARGVLVVNAPTSNIHSAAEHALALLLAASRQIAEADASLRAHIWK
: .* *: * : . . :.** :: *:* ::: * :
Mb0749c|M.bovis_AF2122/97 RDGTIPYQRFRGAEIAGLTAGLVGLGAVGRAVRWRLSGLGLRVIAHDPYR
Rv0728c|M.tuberculosis_H37Rv RDGTIPYQRFRGAEIAGLTAGLVGLGAVGRAVRWRLSGLGLRVIAHDPYR
MMAR_1065|M.marinum_M HDGTIPYQRFRGWEIAGRTAGLVGLGAVGRATAWRLTGLGLRVIAHDPYC
MUL_0823|M.ulcerans_Agy99 RDGTIPYQRFRGWEIAGRTAGLVGLGAVGRATAWRLTGLGLWVIAHDPYC
MAV_4438|M.avium_104 RDGSIPYQRFRGGEIAGLTAGLVGLGAVGRATRWRLSGLGLRVIAHDPYH
TH_3809|M.thermoresistible__bu -----DRLVLRGSQLQHKTMGIVGLGAIGRRLVQLVAPFGMKVLAFDPYV
Mflv_0334|M.gilvum_PYR-GCK PSDPTLGESV--RDLGGCTVGLVGYGNIAKQVERILLAIGAE-VVHTSTR
MSMEG_0604|M.smegmatis_MC2_155 PTDPTLGDTV--RDIGGCTVGLVGYGNVAKRVERIVLAMGAEQVLHTSTR
MAB_3304c|M.abscessus_ATCC_199 RSSFNG------TEIFGKTVGVVGLGRIGQLFAQRLAAFGTHIVAYDPYV
Mvan_2128|M.vanbaalenii_PYR-1 RSSFSG------TEIFGKTVGVVGLGRIGQLVAQRLAAFGAHITAYDPYV
MLBr_01692|M.leprae_Br4923 RSSFSG------TEIFGKTVGVVGLGRIGQLVAARIAAFGAHVIAYDPYV
:: * *:** * :.: : :* . .
Mb0749c|M.bovis_AF2122/97 DDAG--------HSLDELLAEADIVSMHAAVTDDTIGMIGAQQFAAMRDG
Rv0728c|M.tuberculosis_H37Rv DDAG--------HSLDELLAEADIVSMHAAVTDDTIGMIGAQQFAAMRDG
MMAR_1065|M.marinum_M ADAR--------HGLAELLAESDIVSLHAPVTDDTVGMIGAQQFASMRDG
MUL_0823|M.ulcerans_Agy99 ADAR--------HGLAELLAESDIVSLHAPVTDDTVGMIGAQQFASMRDG
MAV_4438|M.avium_104 PEAR--------HGLDELLAESDVVSLHAPVTDETTGMIGAEQFAAMRDG
TH_3809|M.thermoresistible__bu DEKTAEALGARSVSLEELLGCSDFVQLTCPLTAETEGLIGRAQFAAMKPT
Mflv_0334|M.gilvum_PYR-GCK DDGLPGWR-----TLPDLLSESDIVSLHLPLTGATEGLLGRKALAQMKTD
MSMEG_0604|M.smegmatis_MC2_155 DTGHPRWR-----NLPDLLAASDIVSLHLPLTDTSRGLLGPEAIAAMKPG
MAB_3304c|M.abscessus_ATCC_199 SAARAAQLGIELLTLDELLGRADFISVHLPKTPETAGLIGKEALAKTKPG
Mvan_2128|M.vanbaalenii_PYR-1 SHARAAQLGIELLTLDELLGRADFISVHLPKTKETAGLIGKEALAKTKPG
MLBr_01692|M.leprae_Br4923 APARAAQLGIELMSFDDLLARADFISVHLPKTPETAGLIDKEALAKTKPG
: :**. :*.:.: . * : *::. :* :
Mb0749c|M.bovis_AF2122/97 AVFLNTARSQLHDTDALVDALRGGKLAAAGLDHFTGEWLPTDHPLVSMPN
Rv0728c|M.tuberculosis_H37Rv AVFLNTARSQLRDTDALVDALRGGKLAAAGLDHFTGEWLPTDHPLVSMPN
MMAR_1065|M.marinum_M AVFLNTARAQLHDTDALVQALRAGKVAAAGLDHFVGEWLPTDHPLASMPN
MUL_0823|M.ulcerans_Agy99 AVFLNTARAQLHDTDALVQALRAGKVAAAGLDHFVGEWLPTDHPLASMPN
MAV_4438|M.avium_104 VVFLNTARAQLHDTDALVEALRAGKVAAAGLDHFVGEWLPTDHPLVGMPN
TH_3809|M.thermoresistible__bu AFFVTTARGRVHDEAALYEALTCGEIAGAGLDVFHDEPPRADHPLLTLDN
Mflv_0334|M.gilvum_PYR-GCK AVLVNTSRGAIVDEAALVDALRAGTLAAAGLDVFAVEPVPSDNPLLGLPN
MSMEG_0604|M.smegmatis_MC2_155 AVLVNTARGPIVDEAALIEALRGGRLAAAGLDVFDTEPLPADHPLLGLDN
MAB_3304c|M.abscessus_ATCC_199 VIIVNAARGGLIDEAALADAIRSGHVRGAGLDVFSTEPCT-DSPLFELDQ
Mvan_2128|M.vanbaalenii_PYR-1 VIIVNAARGGLIDEAALADAINSGHVRGAGLDVFSTEPCT-DSPLFELPQ
MLBr_01692|M.leprae_Br4923 VIIVNAARGGLVDEVALADAVRSGHVRAAGLDVFATEPCT-DSPLFELSQ
..::.::*. : * ** :*: * : .**** * * * ** : :
Mb0749c|M.bovis_AF2122/97 VVLTPHIGGATWNTEARQARMVADDLGALLSGNRPAHVVNPEVLGS----
Rv0728c|M.tuberculosis_H37Rv VVLTPHIGGATWNTEARQARMVADDLGALLSGNRPAHVVNPEVLGS----
MMAR_1065|M.marinum_M VVLTPHIGGATWNTEARQAQMVADGLQALLSGTRPTHILNPEVLAR----
MUL_0823|M.ulcerans_Agy99 VVLTPHIGGATWNTEARQAQMVADGLQALLSGTRPTHILNPEVLAR----
MAV_4438|M.avium_104 VVLTPHIGGATWNTEARQAQLVADDLEALLAGATPAHIVNPEVLAR----
TH_3809|M.thermoresistible__bu VVATPHIAGITAEAARDIAVATADQWQTIFAGGVPPRLLNPEVWPRYSAR
Mflv_0334|M.gilvum_PYR-GCK VVLTPHVTWYTADTMRRYLEFALDNCERLRDGRALAHVVNDPSG------
MSMEG_0604|M.smegmatis_MC2_155 VVLTPHVTWYTADTMRRYLSIGVENCRRIRDGEPLAHLVNEAR-------
MAB_3304c|M.abscessus_ATCC_199 VVVTPHLGASTSEAQDRAGTDVAASVQLALAGEFVPDAVNVGGGVVSEEV
Mvan_2128|M.vanbaalenii_PYR-1 VVVTPHLGASTVEAQDRAGTDVAASVKLALAGEFVPDAVNVGGGAVGEEV
MLBr_01692|M.leprae_Br4923 VVVTPHLGASTAEAQDRAGTDVAESVRLALAGEFVPDAVNVDGGVVNEEV
** ***: * :: * . :*
Mb0749c|M.bovis_AF2122/97 --------------------------------------------------
Rv0728c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1065|M.marinum_M --------------------------------------------------
MUL_0823|M.ulcerans_Agy99 --------------------------------------------------
MAV_4438|M.avium_104 --------------------------------------------------
TH_3809|M.thermoresistible__bu FAEIFGFAPEPVSRTREREAVGR---------------------------
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3304c|M.abscessus_ATCC_199 APWLEVVRKLGVLIGAVSEQLPTSLSVDVRGELASEDVAVLKLSALRGLF
Mvan_2128|M.vanbaalenii_PYR-1 APWLDLVRKLGLLVGVLSSEPPVSLQVQVQGELASEEVEVLKLSALRGLF
MLBr_01692|M.leprae_Br4923 APWLDLVCKLGVLVAALSDELPASLSVHVRGELASEDVEILRLSALRGLF
Mb0749c|M.bovis_AF2122/97 --------------------------------------------------
Rv0728c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1065|M.marinum_M --------------------------------------------------
MUL_0823|M.ulcerans_Agy99 --------------------------------------------------
MAV_4438|M.avium_104 --------------------------------------------------
TH_3809|M.thermoresistible__bu --------------------------------------------------
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3304c|M.abscessus_ATCC_199 SSVIEDQVTFVNAPSIAEERGVSAEITTATESPNHRSVVDVRAVYADGSV
Mvan_2128|M.vanbaalenii_PYR-1 SAVIEHPVTFVNAPALASERGVEASITTATESANHRSVVDVRAVAADGST
MLBr_01692|M.leprae_Br4923 STVIEDAVTFVNAPALAAERGVSAEITTGSESPNHRSVVDVRAVASDGSV
Mb0749c|M.bovis_AF2122/97 --------------------------------------------------
Rv0728c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1065|M.marinum_M --------------------------------------------------
MUL_0823|M.ulcerans_Agy99 --------------------------------------------------
MAV_4438|M.avium_104 --------------------------------------------------
TH_3809|M.thermoresistible__bu --------------------------------------------------
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3304c|M.abscessus_ATCC_199 INVAGTLSGPQLVQKIVQINGRNFDLRAEGVNLVINYADVPGALGKIGTV
Mvan_2128|M.vanbaalenii_PYR-1 VNVAGTLTGPQLVEKIVQINGRNLELRAEGVNLIINYDDQPGALGKIGTL
MLBr_01692|M.leprae_Br4923 VNIAGTLSGPQLVQKIVQVNGRNFDLRAQGMNLVIRYVDQPGALGKIGTL
Mb0749c|M.bovis_AF2122/97 --------------------------------------------------
Rv0728c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1065|M.marinum_M --------------------------------------------------
MUL_0823|M.ulcerans_Agy99 --------------------------------------------------
MAV_4438|M.avium_104 --------------------------------------------------
TH_3809|M.thermoresistible__bu --------------------------------------------------
Mflv_0334|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0604|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3304c|M.abscessus_ATCC_199 LGGAEVNIQAAQLSQDASGAAATIILRIDRTAPDAVLDEIRAAVGATTLE
Mvan_2128|M.vanbaalenii_PYR-1 LGGAAVNILAAQLSQDADGIGATVMLRLDREVPGEVLAAIGRDVNAVTLE
MLBr_01692|M.leprae_Br4923 LGAAGVNIQAAQLSEDTEGPGATILLRLDQDVPGDVRSAIVAAVSANKLE
Mb0749c|M.bovis_AF2122/97 -----
Rv0728c|M.tuberculosis_H37Rv -----
MMAR_1065|M.marinum_M -----
MUL_0823|M.ulcerans_Agy99 -----
MAV_4438|M.avium_104 -----
TH_3809|M.thermoresistible__bu -----
Mflv_0334|M.gilvum_PYR-GCK -----
MSMEG_0604|M.smegmatis_MC2_155 -----
MAB_3304c|M.abscessus_ATCC_199 LVDLS
Mvan_2128|M.vanbaalenii_PYR-1 VVDLT
MLBr_01692|M.leprae_Br4923 VVNLS