For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DRYTVISADCHAGADLLDYRDYLDPQYRDEFDGWAKTYVNPFGDLSEPDAERNWNSARRNCELDTEGIAG EVIYPNTVPPFFPHASLAAPPPETARELELRWAGLRAHNRWLADFCSLSPERRAGVGQILLGDLDEAVAE VAQIAKLGLRGGVLLPGIAPGTGIPPLYAEHWEPLWAACAEAGVVVNHHGGNAGPSPTDGWGSSFAVWVY ETHWWAHRALWHLIFSGALDRHPDLTVVFTEQGAGWIPATLASLDVAAARYARANSAIARFAGPTAGSLS LKPSQYWARQCYVGASFMRPVECAERHGIGVEKIMWGSDYPHYEGTAPYTREALRHTFSDVPPDEVAAMV GGNAARVYGFDLDALAPLVDRIGPSVAEVAEPLAAVPADASSTVFEPDPIRTW*
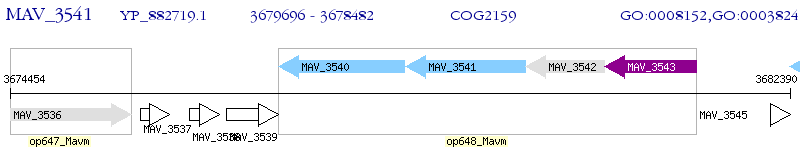
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3541 | - | - | 100% (404) | amidohydrolase |
| M. avium 104 | MAV_3544 | - | 3e-94 | 45.88% (388) | amidohydrolase |
| M. avium 104 | MAV_3540 | - | 2e-74 | 39.35% (399) | amidohydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4324 | - | 2e-21 | 26.52% (313) | amidohydrolase 2 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4820 | - | 0.0 | 86.63% (404) | hypothetical protein MMAR_4820 |
| M. smegmatis MC2 155 | MSMEG_4837 | - | 2e-20 | 28.47% (274) | amidohydrolase 2 |
| M. thermoresistible (build 8) | TH_2277 | - | 3e-21 | 28.57% (308) | amidohydrolase family protein |
| M. ulcerans Agy99 | MUL_0389 | - | 0.0 | 85.15% (404) | hypothetical protein MUL_0389 |
| M. vanbaalenii PYR-1 | Mvan_0901 | - | 7e-25 | 29.06% (320) | amidohydrolase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4820|M.marinum_M --MDRYTVISADCHAGAD-----LLDYREYLEAKYQDEFD---DWAKTFV
MUL_0389|M.ulcerans_Agy99 --MDRYTVISADCHAGAD-----LFDYREYLEAKYQDEFD---DWAKTFV
MAV_3541|M.avium_104 --MDRYTVISADCHAGAD-----LLDYRDYLDPQYRDEFD---GWAKTYV
Mflv_4324|M.gilvum_PYR-GCK MNRDDMILISVDDHIVEP-----PDMFKNHLSKKYLVEAP---RLVHNED
MSMEG_4837|M.smegmatis_MC2_155 MNKDDMILISVDDHIIEP-----PDMFKNHLPAKYRDEAP---RLVHNPD
Mvan_0901|M.vanbaalenii_PYR-1 -MGSSTLIIDTDTHITEP-----PDLWTSRMSKSRWGNLIPEVKWVEEKQ
TH_2277|M.thermoresistible__bu -MSTPTLVYPAEGYGAPKNRRGRAPRDVTELGLPAGTEVFSADNHISVAE
: .: : : :
MMAR_4820|M.marinum_M N----PFGDLSEPDAEHNWDSDR------RNAELDSDGVAGEVIFPNTVP
MUL_0389|M.ulcerans_Agy99 N----PFGDLSEPDAEHNWDSDR------RNAELDSDGVAGEVIFPNTVP
MAV_3541|M.avium_104 N----PFGDLSEPDAERNWNSAR------RNCELDTEGIAGEVIYPNTVP
Mflv_4324|M.gilvum_PYR-GCK GSDTWQFRDVVIPNVALNAVAGR------PKEEYGLEPQGLDEIRPGCWQ
MSMEG_4837|M.smegmatis_MC2_155 GSDTWQFRDTVIPNVALNAVAGR------PKEEYGLEPQGLDEIRKGCYD
Mvan_0901|M.vanbaalenii_PYR-1 GEYWCMDGQPVFTVGTCIMVPGEDGKP-VRSPRFPDYADRFSSMHPSAYD
TH_2277|M.thermoresistible__bu DIFYERFPDDLKGAAPRIWYEDGAYMVGMKGKSWVGGDFGRVLMQYDDLA
. : : .
MMAR_4820|M.marinum_M P----------------FFPQGSLAAPPPSTAR---DLELRWAGLRAHNR
MUL_0389|M.ulcerans_Agy99 P----------------FFPQGSLAAPAPSTAR---DLELRWAGLRAHNR
MAV_3541|M.avium_104 P----------------FFPHASLAAPPPETAR---ELELRWAGLRAHNR
Mflv_4324|M.gilvum_PYR-GCK VDERVKDMNAGGILGSMCFPSFPGFAGRLFATE---DPEFSLALVQAYND
MSMEG_4837|M.smegmatis_MC2_155 PNERVKDMNAGGVLATMNFPSFPGFAARLFATD---DEEFSLALVQAYND
Mvan_0901|M.vanbaalenii_PYR-1 AKARLEVMDAYGIQAAAIFPNLGFVGPNIFAAAGPDALEFQTAALQAYND
TH_2277|M.thermoresistible__bu GAATNNIEARVQELAEDGITRELAFPNAVLALFHYPDKALRERVFRIYNE
:. : : .: :*
MMAR_4820|M.marinum_M W-LADFCSLSPERRAGVGQILLGDVDEAVAEVAQIAKLGLRGGVLLPGV-
MUL_0389|M.ulcerans_Agy99 W-LADFCSLSPERRAGVGQILLGDVDEAVAEVAQIAKLGLPGGVLLPGV-
MAV_3541|M.avium_104 W-LADFCSLSPERRAGVGQILLGDLDEAVAEVAQIAKLGLRGGVLLPGI-
Mflv_4324|M.gilvum_PYR-GCK WHVEEWCGAYPARFIPMTLPVIWDPEACAKEIRRNAARGVHS-LTFSEN-
MSMEG_4837|M.smegmatis_MC2_155 WHIDEWCASNPGRFIPMAIPAIWNPGLAAAEVRRVAEKGVHS-LTFTEN-
Mvan_0901|M.vanbaalenii_PYR-1 F-LLDWSSIAPDRLLSLALIPYWDVDAAVAEIERCAAAGHKG-LVSTGK-
TH_2277|M.thermoresistible__bu H-IADLQDRSNGHFYGVGLINWWDPKGTRSTLEQLKSLGLKTFLLPLNPG
: : : : : : : * :
MMAR_4820|M.marinum_M APGTGIPALY-AEHWEPLWAACAQAGLVINHHGG--------NAGPSPID
MUL_0389|M.ulcerans_Agy99 APGTGIPALY-AEHWEPLWAACAQAGLVINHHGG--------NAGPSPID
MAV_3541|M.avium_104 APGTGIPPLY-AEHWEPLWAACAEAGVVVNHHGG--------NAGPSPTD
Mflv_4324|M.gilvum_PYR-GCK PSAMGYPSFHDFDHWKPMWDALVDTDTVLNVHIG--------SSGRLAIT
MSMEG_4837|M.smegmatis_MC2_155 PSTLGYPSFHDLEYWRPLWQALVDTETVMNVHIG--------SSGKLAIT
Mvan_0901|M.vanbaalenii_PYR-1 PHEHGYKLLA-DRYWDPMWAAAQDANLSISFHVGGGNLSRHLNDERTSVE
TH_2277|M.thermoresistible__bu KDDDGNIYDYGSRAMDPVWDEIEEAGLPVTHHIG---------ETPPKTP
* *:* :: :. * *
MMAR_4820|M.marinum_M GWGSSLAVWVYESHWWAHRALWHLIFSGVLDRYPDLTVVLTEQGTGWIPA
MUL_0389|M.ulcerans_Agy99 GWGSSLAVLVYESHWWAHRALLHLIFSGVLDRYPDLTVVLTEQGTGWIPA
MAV_3541|M.avium_104 GWGSSFAVWVYETHWWAHRALWHLIFSGALDRHPDLTVVFTEQGAGWIPA
Mflv_4324|M.gilvum_PYR-GCK APDAPMDVMITLQPMNIVQAAADLLWSRPIKEYPDLKIALSEGGTGWIPY
MSMEG_4837|M.smegmatis_MC2_155 APDAPMDVMITLQPMNIVQAAADLLWSAPIKEFPTLKIALSEGGTGWIPY
Mvan_0901|M.vanbaalenii_PYR-1 GWRATLARLTTSFFLESGITLADLLMSGVLARYPKLRFVSVESAVGWIPF
TH_2277|M.thermoresistible__bu CEHNSVVVGMMVNVDSFREQFSKYVFSGILDRHPGLRIGWFEGGIAWVPT
.. . : * : ..* * . * . .*:*
MMAR_4820|M.marinum_M TLDSLDVAAARYRRPNSAIARFAGPTAGSLPLQPSQYWARQCYVGASFLR
MUL_0389|M.ulcerans_Agy99 TLDSLDVAAARYRRPNSAIARFAGPTAGSLPLQPSQYWARQCYVGASFLR
MAV_3541|M.avium_104 TLASLDVAAARYARANSAIARFAGPTAGSLSLKPSQYWARQCYVGASFMR
Mflv_4324|M.gilvum_PYR-GCK FLERVDRTYEMHS------TWTGQDFKGKLPSEVFREHFLTCFIAD----
MSMEG_4837|M.smegmatis_MC2_155 FLDRVDRTFEMHS------TWTHQNFGGKLPSEVFREHFMTCFISD----
Mvan_0901|M.vanbaalenii_PYR-1 LLESLDFHYKKYEP------WLERPEFAGNDMLPSDYFHRQVYANY-WYE
TH_2277|M.thermoresistible__bu ALQDAEHLLASYR-----------HMFNHELQHDIRHYWDNHMSASFMVD
* : :
MMAR_4820|M.marinum_M PVECAQRHRIGVDRIMWGSDYPHMEGTAPYSREALRHTFS----DVDPDE
MUL_0389|M.ulcerans_Agy99 PVECAQRHRIGVDRIMWASDYPHMEGTAPYSREALRHTFS----DVDPDE
MAV_3541|M.avium_104 PVECAERHGIGVEKIMWGSDYPHYEGTAPYTREALRHTFS----DVPPDE
Mflv_4324|M.gilvum_PYR-GCK PVGVATRHQIGVDNICWEADYPHSDSMWPGAPEQLDEVLKA--NNVPDDE
MSMEG_4837|M.smegmatis_MC2_155 PVGVKNRHMIGIDNICWEMDYPHSDSMWPGAPEELSAVFDT--YDVPDDE
Mvan_0901|M.vanbaalenii_PYR-1 DLQPWHVAAIGEDNLMFETDYPHQTCLD---KEEIQHSINEGLSGVSETT
TH_2277|M.thermoresistible__bu PLGLELIDRIGVDNVFWSSDYPHNESTYGYSEKSLAAVVQA----VGPEN
: ** :.: : **** : : .. *
MMAR_4820|M.marinum_M VAAMVGGNAAAVYGFDLQALAPLAARIGPTVTEVAEPLAAIPADAS----
MUL_0389|M.ulcerans_Agy99 VAAMVGGNAAAVYGFDLQALAPLAARIGPTVTEVAEPLAAIPADAS----
MAV_3541|M.avium_104 VAAMVGGNAARVYGFDLDALAPLVDRIGPSVAEVAEPLAAVPADAS----
Mflv_4324|M.gilvum_PYR-GCK INKMTYENAMRWYHWDPFTHIAKEQATVGALRKAAEGHDVSIQALSHHKE
MSMEG_4837|M.smegmatis_MC2_155 INKITHENAMRLYHFDPFAHVPKEQATVGALRKAAEGHDVSIRALS--KK
Mvan_0901|M.vanbaalenii_PYR-1 KEKILWRNAASLFNMDIAKLEKLT--------------------------
TH_2277|M.thermoresistible__bu AVKIVSTNIQRFLGIAE---------------------------------
: *
MMAR_4820|M.marinum_M ------STAFEPDPIRAW-----
MUL_0389|M.ulcerans_Agy99 ------STAFEPDPIRAW-----
MAV_3541|M.avium_104 ------STVFEPDPIRTW-----
Mflv_4324|M.gilvum_PYR-GCK GERGGGAGHFDALNAAARGNSGA
MSMEG_4837|M.smegmatis_MC2_155 DKTGASFADFQANAKAVSGARD-
Mvan_0901|M.vanbaalenii_PYR-1 -----------------------
TH_2277|M.thermoresistible__bu -----------------------