For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASTFTDSKSELMRRAAEQLTALQGRDAAESPLTTRQKLALTCRALFDAGHDSGLAGQITARAEADGTYYT QRLGLGFDEITDGNLLLVDEDLNVLEGDGMANPANRFHSWIYRARPDVQCIVHTHAFHVAALSMLEVPLV VSHMDTTPLYDDCAFLADWPGVPVGNEEGEIITAALGDKKAVLLAHHGHVIAGASIEEACSLGILIERAA KLQLAAMAAGTVKQLPEELAREAHDWTLSPQRSRANFAYYARRALARHPDALTS*
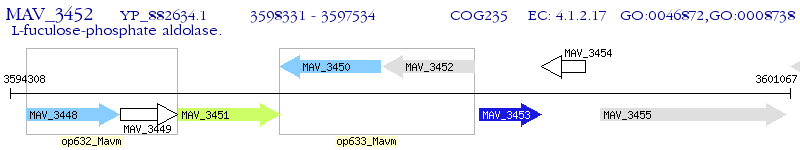
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_3452 | - | - | 100% (265) | hypothetical protein MAV_3452 |
| M. avium 104 | MAV_2994 | - | 9e-18 | 27.51% (189) | putative class II aldolase |
| M. avium 104 | MAV_4439 | - | 1e-08 | 25.87% (201) | L-fuculose-phosphate aldolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0748c | fucA | 1e-09 | 27.57% (185) | L-fuculose-phosphate aldolase |
| M. gilvum PYR-GCK | Mflv_0622 | - | 5e-11 | 29.77% (215) | class II aldolase/adducin family protein |
| M. tuberculosis H37Rv | Rv0727c | fucA | 1e-09 | 27.57% (185) | L-fuculose-phosphate aldolase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1048c | - | 1e-120 | 79.17% (264) | class II aldolase/adducin domain protein |
| M. marinum M | MMAR_2153 | - | 1e-127 | 82.64% (265) | hypothetical protein MMAR_2153 |
| M. smegmatis MC2 155 | MSMEG_6863 | - | 3e-11 | 26.73% (202) | hypothetical protein MSMEG_6863 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3144 | araD | 1e-16 | 25.93% (189) | ribulose-5-phosphate 4-epimerase, AraD |
| M. vanbaalenii PYR-1 | Mvan_0451 | - | 1e-123 | 81.75% (263) | hypothetical protein Mvan_0451 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0748c|M.bovis_AF2122/97 ------MNFVD-----DPESAVLAAAKD----------------------
Rv0727c|M.tuberculosis_H37Rv ------MNFVD-----APESAVLAAAKD----------------------
MAV_3452|M.avium_104 -----MASTFT-----DSKSELMRRAAEQLTALQGRDAAESPLTTRQKLA
MMAR_2153|M.marinum_M -----MATTFS-----DSKSELMRRAQERLDAQT----PESQLTTRQKVA
MAB_1048c|M.abscessus_ATCC_199 -----MASTLH-----DTKSELMDRAERSMQTHF----SDSQWTTRQKVA
Mvan_0451|M.vanbaalenii_PYR-1 -----MTSTMH-----DSKSDLMRRALDGLSRNV----TESSLTLREKLA
Mflv_0622|M.gilvum_PYR-GCK -----MSSTPSG-AGLDTERHAIA--------------------------
MUL_3144|M.ulcerans_Agy99 MPKGSFATIFEPGWRPSMNAPDARQGGLQVWAPSVIPPIGVELSDAQALA
MSMEG_6863|M.smegmatis_MC2_155 -------MKME------QRSPAPTEAELR-----------------RELA
.
Mb0748c|M.bovis_AF2122/97 -----MLRRGLVEGTAGNISARR-SDGNVVITP----SSVDYAEMLLHDL
Rv0727c|M.tuberculosis_H37Rv -----MLRRGLVEGTAGNISARR-SDGNVVITP----SSVDYAEMLLHDL
MAV_3452|M.avium_104 LTCRALFDAGHDSGLAGQITARAEADGTYYTQR----LGLGFDEITDGNL
MMAR_2153|M.marinum_M LTCRVLFDAGHDSGLAGQITARAQRPGTYYTQQ----LGLGFDEITEDNL
MAB_1048c|M.abscessus_ATCC_199 LTCRALFDRGHDSGLAGQITARAEEPGTFYTQR----LGLGFDEITEENL
Mvan_0451|M.vanbaalenii_PYR-1 VTCRALFDAGHDSGLAGQITARAEAPGTYYTQR----LGLGFDEITAGNL
Mflv_0622|M.gilvum_PYR-GCK LACRVLAARDLAPGILGHISLRVDRDRLLIRCRGPRERGLAFTSAEDIRM
MUL_3144|M.ulcerans_Agy99 VAFRHLAGIGFAENMAGHITWQPDGQTDMLVNP----WGLWWQELTASDI
MSMEG_6863|M.smegmatis_MC2_155 AVYRLVAHYRMTDLVFTHISVRLPGNDHHFLIN---PYGLLFEEITASSL
: :*: : .: : . :
Mb0748c|M.bovis_AF2122/97 VLVD-AGGAVLHAKDGRSPSTELNLHLACYRAFDDIGSVIHSHP--VWAT
Rv0727c|M.tuberculosis_H37Rv VLVD-AGGAVLHAKDGRSPSTELNLHLACYRAFDDIGSVIHSHP--VWAT
MAV_3452|M.avium_104 LLVD-EDLNVLEGDGMANPANRF--HSWIYRARPDVQCIVHTHA--FHVA
MMAR_2153|M.marinum_M LLVD-EDLNVLEGSAMANPANRF--HSWIYRERPDVQCIVHTHG--FHTA
MAB_1048c|M.abscessus_ATCC_199 LLVD-EDLNVLEGSGMANPANRF--HSWIYRARPDVQCIVHTHP--FHVA
Mvan_0451|M.vanbaalenii_PYR-1 LLVD-EDLKVLDGEGMANPANRF--HSWIYRARPDVNCIVHTHP--FHVA
Mflv_0622|M.gilvum_PYR-GCK VTLDGTAGGKGELDDGYQPPNELPLHTEVLRTRRDVNAVVHAHPEAVVAA
MUL_3144|M.ulcerans_Agy99 CTVD-ADARVVKGRWDVTPAIHI--HTQLHRVRDDARVVIHNHP--YYVC
MSMEG_6863|M.smegmatis_MC2_155 VRVDLNGNVIGETPYCVNP-AGFVIHSAIHAAREDAACVLHTHT---LAG
:* . * : * * ::* * .
Mb0748c|M.bovis_AF2122/97 MFAVAHEPIPACIDEFAIYCGGDVRCTEYAA--SGTP-EVGRNAVRALEG
Rv0727c|M.tuberculosis_H37Rv MFAVAHEPIPACIDEFAIYCGGDVRCTEYAA--SGTP-EVGRNAVRALEG
MAV_3452|M.avium_104 ALSMLEVPLVVSHMDTTPLYDDCAFLADWPG--VPVGNEEGEIITAALGD
MMAR_2153|M.marinum_M ALSMLKVPLIVSHMDTTPLYDDCAFLADWPG--VPVGNEEGEIISAALGN
MAB_1048c|M.abscessus_ATCC_199 ALSMLETPLIVSQMDIAPLYDDCAFLPDWPG--VPVGNEEGEIISAALGD
Mvan_0451|M.vanbaalenii_PYR-1 ALSMLEVPLVVSQMDIAPLYGDCAFLPDWPG--VPVGNEEGEIISAALGD
Mflv_0622|M.gilvum_PYR-GCK DLAGLAVRPIVGAFDIPGFRLAAAGVPVYRRGVLVRNRQLAQEMVAAMAD
MUL_3144|M.ulcerans_Agy99 VLAALGRLPDLVHQTGSLFLDDMCLVESYDG--EIDSAARAADLAAHIGS
MSMEG_6863|M.smegmatis_MC2_155 CAVAASASGLLPVNQISMEFFNRVGYHDYEG--VALNLDEQKRLVEDLGT
. : :
Mb0748c|M.bovis_AF2122/97 RAAALIANHGLVAVGPRPDQVLRVTALVERTAQIVWGARALG-GPVPIPE
Rv0727c|M.tuberculosis_H37Rv RAAALIANHGLVAVGPRPDQVLRVTALVERTAQIVWGARALG-GPVPIPE
MAV_3452|M.avium_104 KKAVLLAHHGHVIAGASIEEACSLGILIERAAKLQLAAMAAG-TVKQLPE
MMAR_2153|M.marinum_M KKAILLAHHGQLIAGISIEEACTLAVLIERAAKLQLAAMSAG-TIAELPD
MAB_1048c|M.abscessus_ATCC_199 KKAILLAHHGHVVAGASVEESCSLAVLIERGAKLQLAAMAAG-TIAPLPD
Mvan_0451|M.vanbaalenii_PYR-1 KKAILLAHHGHVVAGASVEEACSLAMLVERGAKLQLAAMAAG-TIAPLPD
Mflv_0622|M.gilvum_PYR-GCK RPVVVLRAHGLTSAADTVERAVLQAISVDTISRLSLQIASAGGTLADLPD
MUL_3144|M.ulcerans_Agy99 ANLTILANHGVIATGRNLAQAVYRAASIEQVCKLAYDVMLTGAEPTQMKW
MSMEG_6863|M.smegmatis_MC2_155 HDALILRNHGLLTVGATPARAFLRMYYLNKACEIQIAASQLGELVIPDPR
:: ** .. . :: ..: *
Mb0748c|M.bovis_AF2122/97 DVCRN------------------FTGVYGYLRANPL--------------
Rv0727c|M.tuberculosis_H37Rv DVCRN------------------FTGVYGYLRANPL--------------
MAV_3452|M.avium_104 ELAREAHDWT--------LSPQRSRANFAYYARRALARHPDALTS-----
MMAR_2153|M.marinum_M QLAREAHDWT--------LTPSRSRANFAYYARRALARHPDALSS-----
MAB_1048c|M.abscessus_ATCC_199 RLAREAHDWT--------LTPKRSIANFAYYARTALAHHPETLTLPTTQG
Mvan_0451|M.vanbaalenii_PYR-1 RLAREAHDWT--------LTPKRSQANFAYYARRALANHPDALDR-----
Mflv_0622|M.gilvum_PYR-GCK ADAAELPDLGN-----AFNETIAWRHELARLETHGLSCHPSEKRSS----
MUL_3144|M.ulcerans_Agy99 SDMAGMRSS---------LIERAADVYWAGAARMTIKADPAVLS------
MSMEG_6863|M.smegmatis_MC2_155 IIEHAAQQLVGDAAGDDYTDDVGYDLAWAAMVRMLDRTQPDYRD------
.
Mb0748c|M.bovis_AF2122/97 --
Rv0727c|M.tuberculosis_H37Rv --
MAV_3452|M.avium_104 --
MMAR_2153|M.marinum_M --
MAB_1048c|M.abscessus_ATCC_199 NS
Mvan_0451|M.vanbaalenii_PYR-1 --
Mflv_0622|M.gilvum_PYR-GCK --
MUL_3144|M.ulcerans_Agy99 --
MSMEG_6863|M.smegmatis_MC2_155 --