For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKMEQRSPAPTEAELRRELAAVYRLVAHYRMTDLVFTHISVRLPGNDHHFLINPYGLLFEEITASSLVRV DLNGNVIGETPYCVNPAGFVIHSAIHAAREDAACVLHTHTLAGCAVAASASGLLPVNQISMEFFNRVGYH DYEGVALNLDEQKRLVEDLGTHDALILRNHGLLTVGATPARAFLRMYYLNKACEIQIAASQLGELVIPDP RIIEHAAQQLVGDAAGDDYTDDVGYDLAWAAMVRMLDRTQPDYRD
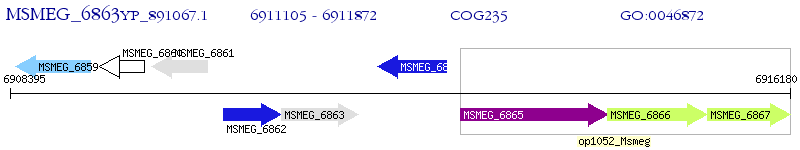
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6863 | - | - | 100% (255) | hypothetical protein MSMEG_6863 |
| M. smegmatis MC2 155 | MSMEG_1714 | - | 2e-06 | 25.44% (169) | L-ribulose-5-phosphate 4-epimerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2912c | - | 5e-33 | 36.52% (230) | aldolase |
| M. marinum M | MMAR_2617 | araD | 5e-15 | 29.32% (191) | ribulose-5-phosphate 4-epimerase, AraD |
| M. avium 104 | MAV_2994 | - | 2e-14 | 28.11% (249) | putative class II aldolase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3144 | araD | 3e-15 | 27.02% (248) | ribulose-5-phosphate 4-epimerase, AraD |
| M. vanbaalenii PYR-1 | Mvan_0451 | - | 3e-10 | 30.82% (159) | hypothetical protein Mvan_0451 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2912c|M.abscessus_ATCC_199 -----MTSGNVVAGEELSAYMVGSADGVQLGPAAAETVTQERERRLRELA
Mvan_0451|M.vanbaalenii_PYR-1 -----MTS---TMHDSKSDLMRRALDGLSR------NVTESSLTLREKLA
MMAR_2617|M.marinum_M -----------------MNAPDARQGGLQVWAPSVIPPIGVELSDAQALA
MUL_3144|M.ulcerans_Agy99 MPKGSFATIFEPGWRPSMNAPDARQGGLQVWAPSVIPPIGVELSDAQALA
MAV_2994|M.avium_104 -----------------MNAPDARRGGLEVWAPSVVPPIGVELSDEQALA
MSMEG_6863|M.smegmatis_MC2_155 -----------------MKMEQRS-------------PAPTEAELRRELA
. **
MAB_2912c|M.abscessus_ATCC_199 AAFRIFGTFGFSEGVAGHITVRDPEFPDTFWVNPFGMNFRHITVSDLIQV
Mvan_0451|M.vanbaalenii_PYR-1 VTCRALFDAGHDSGLAGQITAR-AEAPGTYYTQRLGLGFDEITAGNLLLV
MMAR_2617|M.marinum_M VAFRHLAGIGFAENMAGHITWQ-PDGQTDMLVNPWGLWWQELTASDICTV
MUL_3144|M.ulcerans_Agy99 VAFRHLAGIGFAENMAGHITWQ-PDGQTDMLVNPWGLWWQELTASDICTV
MAV_2994|M.avium_104 VAFRHLAGIGFAENMAGHITWQ-RDGRTDMFVNPWGLWWQEITASDICVV
MSMEG_6863|M.smegmatis_MC2_155 AVYRLVAHYRMTDLVFTHISVRLPGNDHHFLINPYGLLFEEITASSLVRV
.. * . . : :*: : : *: : .:*...: *
MAB_2912c|M.abscessus_ATCC_199 DHDGNVITGS-RPVNRAAFCIHSEVHKARPDVIAAAHAHSVHGKAFSSLG
Mvan_0451|M.vanbaalenii_PYR-1 DEDLKVLDGE-GMAN-PANRFHSWIYRARPDVNCIVHTHPFHVAALSMLE
MMAR_2617|M.marinum_M DADARVVKGR-WDVT-PAIHIHTQLHRVRDDARVVIHNHPYYVCVLAALG
MUL_3144|M.ulcerans_Agy99 DADARVVKGR-WDVT-PAIHIHTQLHRVRDDARVVIHNHPYYVCVLAALG
MAV_2994|M.avium_104 DGDARVVRGR-WDVT-PAIHIHTELHRVRDDARVVIHNHPYYVCVLAALG
MSMEG_6863|M.smegmatis_MC2_155 DLNGNVIGETPYCVNPAGFVIHSAIHAAREDAACVLHTHTLAGCAVAASA
* : .*: .. .. :*: :: .* *. * *. ..:
MAB_2912c|M.abscessus_ATCC_199 QPLLPITQDACAFYQDHAVYTDYRGVVGDLEEGRAIGSALGPAKAVILQN
Mvan_0451|M.vanbaalenii_PYR-1 VPLVVSQMDIAPLYGDCAFLPDWPGVPVGNEEGEIISAALGDKKAILLAH
MMAR_2617|M.marinum_M RLPDLVHQTGSLFLDDMCLVESYDGEIDSAARAADLAAHIGSANLTILAN
MUL_3144|M.ulcerans_Agy99 RLPDLVHQTGSLFLDDMCLVESYDGEIDSAARAADLAAHIGSANLTILAN
MAV_2994|M.avium_104 TLPELVHQTGSLFLDDLCLVDTYDGEIDTPARAADLAARIGGANLTILAN
MSMEG_6863|M.smegmatis_MC2_155 SGLLPVNQISMEFFNRVG-YHDYEGVALNLDEQKRLVEDLGTHDALILRN
: : * . : :* . :* :
MAB_2912c|M.abscessus_ATCC_199 HGLLTVGHSVAEAAWWFITMERSCQAQLLAMAAGEPRTIDHDTAVSVYNQ
Mvan_0451|M.vanbaalenii_PYR-1 HGHVVAGASVEEACSLAMLVERGAKLQLAAMAAGTIAPLPDRLAREAHDW
MMAR_2617|M.marinum_M HGVIATGRNLAQAVYRAASIERVCKLAYDVMLTGAEPTQMKWSDMAAMRS
MUL_3144|M.ulcerans_Agy99 HGVIATGRNLAQAVYRAASIEQVCKLAYDVMLTGAEPTQMKWSDMAGMRS
MAV_2994|M.avium_104 HGVIATGRNLPEAVYRAASIERVCKLAYDVMLTGREPVSMNWSDMAGMQR
MSMEG_6863|M.smegmatis_MC2_155 HGLLTVGATPARAFLRMYYLNKACEIQIAASQLGELVIPDPRIIEHAAQQ
** :..* . .* ::: .: . *
MAB_2912c|M.abscessus_ATCC_199 IGTPVAGW---------FQFQPLWDDVIRTAPEALH-
Mvan_0451|M.vanbaalenii_PYR-1 TLTPKRSQ---------ANFAYYARRALANHPDALDR
MMAR_2617|M.marinum_M SLIERAAD---------VYWAGAARMTIKADPAVLS-
MUL_3144|M.ulcerans_Agy99 SLIERAAD---------VYWAGAARMTIKADPAVLS-
MAV_2994|M.avium_104 SLIERAAD---------VYWAGAARMTIKADPDVLQ-
MSMEG_6863|M.smegmatis_MC2_155 LVGDAAGDDYTDDVGYDLAWAAMVRMLDRTQPDYRD-
. : *