For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASTLHDTKSELMDRAERSMQTHFSDSQWTTRQKVALTCRALFDRGHDSGLAGQITARAEEPGTFYTQRLG LGFDEITEENLLLVDEDLNVLEGSGMANPANRFHSWIYRARPDVQCIVHTHPFHVAALSMLETPLIVSQM DIAPLYDDCAFLPDWPGVPVGNEEGEIISAALGDKKAILLAHHGHVVAGASVEESCSLAVLIERGAKLQL AAMAAGTIAPLPDRLAREAHDWTLTPKRSIANFAYYARTALAHHPETLTLPTTQGNS*
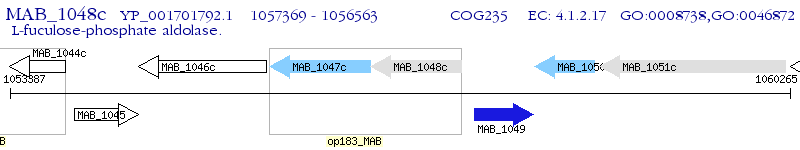
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1048c | - | - | 100% (268) | class II aldolase/adducin domain protein |
| M. abscessus ATCC 19977 | MAB_2912c | - | 1e-28 | 39.47% (190) | aldolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0748c | fucA | 4e-11 | 25.76% (198) | L-fuculose-phosphate aldolase |
| M. gilvum PYR-GCK | Mflv_0622 | - | 2e-13 | 30.73% (218) | class II aldolase/adducin family protein |
| M. tuberculosis H37Rv | Rv0727c | fucA | 4e-11 | 25.76% (198) | L-fuculose-phosphate aldolase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2153 | - | 1e-120 | 80.00% (260) | hypothetical protein MMAR_2153 |
| M. avium 104 | MAV_3452 | - | 1e-120 | 79.17% (264) | hypothetical protein MAV_3452 |
| M. smegmatis MC2 155 | MSMEG_6863 | - | 4e-11 | 28.40% (162) | hypothetical protein MSMEG_6863 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3144 | araD | 5e-16 | 26.88% (186) | ribulose-5-phosphate 4-epimerase, AraD |
| M. vanbaalenii PYR-1 | Mvan_0451 | - | 1e-127 | 84.17% (259) | hypothetical protein Mvan_0451 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0748c|M.bovis_AF2122/97 ------MNFVD----------DPESAVLAAAKD-----------------
Rv0727c|M.tuberculosis_H37Rv ------MNFVD----------APESAVLAAAKD-----------------
MAB_1048c|M.abscessus_ATCC_199 -----MASTLH----------DTKSELMDRAERSMQTHFS----DSQWTT
Mvan_0451|M.vanbaalenii_PYR-1 -----MTSTMH----------DSKSDLMRRALDGLSRNVT----ESSLTL
MMAR_2153|M.marinum_M -----MATTFS----------DSKSELMRRAQERLDAQT----PESQLTT
MAV_3452|M.avium_104 -----MASTFT----------DSKSELMRRAAEQLTALQGRDAAESPLTT
Mflv_0622|M.gilvum_PYR-GCK -----MSSTPSG------AGLDTERHAIA---------------------
MUL_3144|M.ulcerans_Agy99 MPKGSFATIFEPGWRPSMNAPDARQGGLQVWAPSVIPPIG-----VELSD
MSMEG_6863|M.smegmatis_MC2_155 ---MKMEQRSP----------APTEAELRR--------------------
. :
Mb0748c|M.bovis_AF2122/97 ----------MLRRGLVEGTAGNISARR-SDGNVVITP----SSVDYAEM
Rv0727c|M.tuberculosis_H37Rv ----------MLRRGLVEGTAGNISARR-SDGNVVITP----SSVDYAEM
MAB_1048c|M.abscessus_ATCC_199 RQKVALTCRALFDRGHDSGLAGQITARAEEPGTFYTQR----LGLGFDEI
Mvan_0451|M.vanbaalenii_PYR-1 REKLAVTCRALFDAGHDSGLAGQITARAEAPGTYYTQR----LGLGFDEI
MMAR_2153|M.marinum_M RQKVALTCRVLFDAGHDSGLAGQITARAQRPGTYYTQQ----LGLGFDEI
MAV_3452|M.avium_104 RQKLALTCRALFDAGHDSGLAGQITARAEADGTYYTQR----LGLGFDEI
Mflv_0622|M.gilvum_PYR-GCK -----LACRVLAARDLAPGILGHISLRVDRDRLLIRCRGPRERGLAFTSA
MUL_3144|M.ulcerans_Agy99 AQALAVAFRHLAGIGFAENMAGHITWQPDGQTDMLVNP----WGLWWQEL
MSMEG_6863|M.smegmatis_MC2_155 --ELAAVYRLVAHYRMTDLVFTHISVRLPGNDHHFLIN---PYGLLFEEI
: :*: : .: : .
Mb0748c|M.bovis_AF2122/97 LLHDLVLVD-AGGAVLHAKDGRSPSTELNLHLACYRAFDDIGSVIHSHP-
Rv0727c|M.tuberculosis_H37Rv LLHDLVLVD-AGGAVLHAKDGRSPSTELNLHLACYRAFDDIGSVIHSHP-
MAB_1048c|M.abscessus_ATCC_199 TEENLLLVD-EDLNVLEGSGMANPANRF--HSWIYRARPDVQCIVHTHP-
Mvan_0451|M.vanbaalenii_PYR-1 TAGNLLLVD-EDLKVLDGEGMANPANRF--HSWIYRARPDVNCIVHTHP-
MMAR_2153|M.marinum_M TEDNLLLVD-EDLNVLEGSAMANPANRF--HSWIYRERPDVQCIVHTHG-
MAV_3452|M.avium_104 TDGNLLLVD-EDLNVLEGDGMANPANRF--HSWIYRARPDVQCIVHTHA-
Mflv_0622|M.gilvum_PYR-GCK EDIRMVTLDGTAGGKGELDDGYQPPNELPLHTEVLRTRRDVNAVVHAHPE
MUL_3144|M.ulcerans_Agy99 TASDICTVD-ADARVVKGRWDVTPAIHI--HTQLHRVRDDARVVIHNHP-
MSMEG_6863|M.smegmatis_MC2_155 TASSLVRVDLNGNVIGETPYCVNP-AGFVIHSAIHAAREDAACVLHTHT-
: :* . * : * * ::* *
Mb0748c|M.bovis_AF2122/97 -VWATMFAVAHEPIPACIDEFAIYCGGDVRCTEYAA--SGTP-EVGRNAV
Rv0727c|M.tuberculosis_H37Rv -VWATMFAVAHEPIPACIDEFAIYCGGDVRCTEYAA--SGTP-EVGRNAV
MAB_1048c|M.abscessus_ATCC_199 -FHVAALSMLETPLIVSQMDIAPLYDDCAFLPDWPG--VPVGNEEGEIIS
Mvan_0451|M.vanbaalenii_PYR-1 -FHVAALSMLEVPLVVSQMDIAPLYGDCAFLPDWPG--VPVGNEEGEIIS
MMAR_2153|M.marinum_M -FHTAALSMLKVPLIVSHMDTTPLYDDCAFLADWPG--VPVGNEEGEIIS
MAV_3452|M.avium_104 -FHVAALSMLEVPLVVSHMDTTPLYDDCAFLADWPG--VPVGNEEGEIIT
Mflv_0622|M.gilvum_PYR-GCK AVVAADLAGLAVRPIVGAFDIPGFRLAAAGVPVYRRGVLVRNRQLAQEMV
MUL_3144|M.ulcerans_Agy99 -YYVCVLAALGRLPDLVHQTGSLFLDDMCLVESYDG--EIDSAARAADLA
MSMEG_6863|M.smegmatis_MC2_155 --LAGCAVAASASGLLPVNQISMEFFNRVGYHDYEG--VALNLDEQKRLV
. . :
Mb0748c|M.bovis_AF2122/97 RALEGRAAALIANHGLVAVGPRPDQVLRVTALVERTAQIVWGARALG-GP
Rv0727c|M.tuberculosis_H37Rv RALEGRAAALIANHGLVAVGPRPDQVLRVTALVERTAQIVWGARALG-GP
MAB_1048c|M.abscessus_ATCC_199 AALGDKKAILLAHHGHVVAGASVEESCSLAVLIERGAKLQLAAMAAG-TI
Mvan_0451|M.vanbaalenii_PYR-1 AALGDKKAILLAHHGHVVAGASVEEACSLAMLVERGAKLQLAAMAAG-TI
MMAR_2153|M.marinum_M AALGNKKAILLAHHGQLIAGISIEEACTLAVLIERAAKLQLAAMSAG-TI
MAV_3452|M.avium_104 AALGDKKAVLLAHHGHVIAGASIEEACSLGILIERAAKLQLAAMAAG-TV
Mflv_0622|M.gilvum_PYR-GCK AAMADRPVVVLRAHGLTSAADTVERAVLQAISVDTISRLSLQIASAGGTL
MUL_3144|M.ulcerans_Agy99 AHIGSANLTILANHGVIATGRNLAQAVYRAASIEQVCKLAYDVMLTGAEP
MSMEG_6863|M.smegmatis_MC2_155 EDLGTHDALILRNHGLLTVGATPARAFLRMYYLNKACEIQIAASQLGELV
: :: ** .. . :: ..: *
Mb0748c|M.bovis_AF2122/97 VPIPEDVCRN------------------FTGVYGYLRANPL---------
Rv0727c|M.tuberculosis_H37Rv VPIPEDVCRN------------------FTGVYGYLRANPL---------
MAB_1048c|M.abscessus_ATCC_199 APLPDRLAREAHDWT--------LTPKRSIANFAYYARTALAHHPETLTL
Mvan_0451|M.vanbaalenii_PYR-1 APLPDRLAREAHDWT--------LTPKRSQANFAYYARRALANHPDALDR
MMAR_2153|M.marinum_M AELPDQLAREAHDWT--------LTPSRSRANFAYYARRALARHPDALSS
MAV_3452|M.avium_104 KQLPEELAREAHDWT--------LSPQRSRANFAYYARRALARHPDALTS
Mflv_0622|M.gilvum_PYR-GCK ADLPDADAAELPDLGN-----AFNETIAWRHELARLETHGLSCHPSEKRS
MUL_3144|M.ulcerans_Agy99 TQMKWSDMAGMRSS---------LIERAADVYWAGAARMTIKADPAVLS-
MSMEG_6863|M.smegmatis_MC2_155 IPDPRIIEHAAQQLVGDAAGDDYTDDVGYDLAWAAMVRMLDRTQPDYRD-
.
Mb0748c|M.bovis_AF2122/97 -------
Rv0727c|M.tuberculosis_H37Rv -------
MAB_1048c|M.abscessus_ATCC_199 PTTQGNS
Mvan_0451|M.vanbaalenii_PYR-1 -------
MMAR_2153|M.marinum_M -------
MAV_3452|M.avium_104 -------
Mflv_0622|M.gilvum_PYR-GCK S------
MUL_3144|M.ulcerans_Agy99 -------
MSMEG_6863|M.smegmatis_MC2_155 -------