For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRHLSHGLVELGHDVEVFSGQPYPDLLDPRVRLTEVPSLDLYREPDPFRVPRPSEIRDSIDLLELLTTWT AGFPEPRTFTLRAARLLAGRTADFDVVHDNQSLGTGLLSIAGSGLPVVATVHHPITRDRVLDLAAARWWR KPLVRRWYGFLAMQQEVARHIPDLLTVSSSSAADIISDFGVSPEQLHVVPLGVNTELFKPGSGPRQPGRI IAIASADTPLKGVRTLLHAVARLRTVRDLELRLVAKVEPNGPTHKLIAELGISDIVHITSGLSDAELAAL FASAEVACIPSLYEGFSLPAVEAMASGTPIVASRVGALPEVLGTDGACAELVPPADVDALTRALGELLDS PEKRRSLGRAGRTRAVDVFSWEAVAAQTVRVYERAIARNAAGRAPC
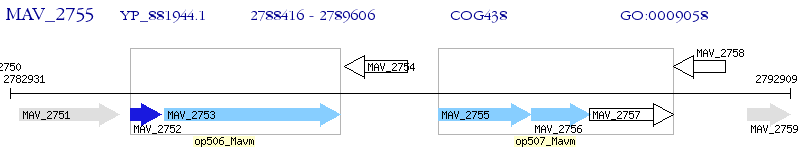
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2755 | - | - | 100% (396) | glycosyl transferase |
| M. avium 104 | MAV_4082 | - | 2e-29 | 34.92% (252) | transferase |
| M. avium 104 | MAV_1357 | - | 3e-25 | 29.74% (343) | glycogen synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0496 | - | 3e-23 | 30.85% (363) | mannosyltransferase |
| M. gilvum PYR-GCK | Mflv_1099 | - | 1e-176 | 80.00% (390) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv0486 | - | 3e-23 | 30.85% (363) | mannosyltransferase |
| M. leprae Br4923 | MLBr_02443 | - | 1e-21 | 29.75% (363) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_1351c | - | 4e-24 | 29.15% (343) | putative glycosyltransferase |
| M. marinum M | MMAR_2925 | - | 0.0 | 86.08% (388) | glycosyltransferase |
| M. smegmatis MC2 155 | MSMEG_6484 | - | 1e-176 | 79.04% (396) | glycosyl transferase |
| M. thermoresistible (build 8) | TH_0088 | - | 1e-178 | 80.57% (386) | glycosyl transferase |
| M. ulcerans Agy99 | MUL_4556 | - | 2e-24 | 31.40% (363) | mannosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5718 | - | 1e-175 | 80.00% (390) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0496|M.bovis_AF2122/97 MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
Rv0486|M.tuberculosis_H37Rv MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
MLBr_02443|M.leprae_Br4923 ------------------------------------------------ML
MUL_4556|M.ulcerans_Agy99 --MRAGAGAAGESCKDDGVRPDDVSR-----------LTADAVVRRVALL
MAV_2755|M.avium_104 --------------------------------------------------
MMAR_2925|M.marinum_M ---------------------------------MRIALLSYRSKTHCGGQ
MSMEG_6484|M.smegmatis_MC2_155 ---------------------------------MRIALLSYRSKTHCGGQ
TH_0088|M.thermoresistible__bu ---------------------------------MRIALLSYRSKTHCGGQ
Mflv_1099|M.gilvum_PYR-GCK ---------------------------------MRIALLSYRSKTHCGGQ
Mvan_5718|M.vanbaalenii_PYR-1 ---------------------------------MRIALLSYRSKTHCGGQ
MAB_1351c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0496|M.bovis_AF2122/97 AVHTSPLAQPGTGDAGGMNVYMLQSALHLARRGIEVEIFTRATASADPPV
Rv0486|M.tuberculosis_H37Rv AVHTSPLAQPGTGDAGGMNVYMLQSALHLARRGIEVEIFTRATASADPPV
MLBr_02443|M.leprae_Br4923 AVHTSPLAQPGIGDAGGMNVYVLQSALHLARRGIEVEIFTRATASADPPI
MUL_4556|M.ulcerans_Agy99 AVHTSPLAQPGTGDAGGMNVYVLQSALHLAKRGIEVEIFTRATASADPPV
MAV_2755|M.avium_104 ---MRHLSHGLVELGHDVEVFSGQPYPDLLDPRVRLTEVPSLDLYREPDP
MMAR_2925|M.marinum_M GVYVRHLSRGLVDLGHDVEVFSGQPYPELLDPRVRLTEVPSLDLYREPDP
MSMEG_6484|M.smegmatis_MC2_155 GVYVRHLSSGLAELGHDVEVFSGQPYPEGLDPRVTLTKVPSLDLYREPDP
TH_0088|M.thermoresistible__bu GVYVRHLSRGLVELGHQVEVFSGQPYPEGLDPRVKLTKVPSLDLYREPDP
Mflv_1099|M.gilvum_PYR-GCK GVYVRHLSRGLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDP
Mvan_5718|M.vanbaalenii_PYR-1 GVYVRHLSRGLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDP
MAB_1351c|M.abscessus_ATCC_199 ---MTRVAMMTREYPPEVYGGAGVHVTELVAQLRTLCDVDVHCMGAPRDN
:: : . : .
Mb0496|M.bovis_AF2122/97 VRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPGYYDI
Rv0486|M.tuberculosis_H37Rv VRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPGYYDI
MLBr_02443|M.leprae_Br4923 VWVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAAHEPGYYDI
MUL_4556|M.ulcerans_Agy99 VRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAAHEPGHYDI
MAV_2755|M.avium_104 FRVPRPSEIRDSID-LLELLTTWTAGFPEPRTFTLRAARLLAGRTADFDV
MMAR_2925|M.marinum_M FRVPHPSEIRDSID-LLELFTMWTAGFPEPRTFSLRVARLLADRVGDFDL
MSMEG_6484|M.smegmatis_MC2_155 FRVPHPREIRDGID-ALELLTMWSAGFPEPRTFTMRAARILAARRDEFDV
TH_0088|M.thermoresistible__bu FRIPRPSEIRDRID-LLELLTTWTAGFPEPKTFTMRMARILAERRDEFDV
Mflv_1099|M.gilvum_PYR-GCK FRIPRPSEIRSSID-LEELLTTWTAGFPEPKTFSMRAARLLAARRGDFDV
Mvan_5718|M.vanbaalenii_PYR-1 FRIPRPSEIKTSID-LEELLTTWTAGFPEPKTFSMRAARLLADRRDDFDV
MAB_1351c|M.abscessus_ATCC_199 AFVHRPDPALDGAN---------------PALTTLSADLVMAAAAAGSTV
: : . :
Mb0496|M.bovis_AF2122/97 VHSHYWLSGQVGWLARDRWAVPLVHTAHT-LAAVKNAALADGDGPEPPLR
Rv0486|M.tuberculosis_H37Rv VHSHYWLSGQVGWLARDRWAVPLVHTAHT-LAAVKNAALADGDGPEPPLR
MLBr_02443|M.leprae_Br4923 VHSHYWLSGQVGWLARDRWAVPLVHTAHT-LAAVKNAALADGDAAEPPLR
MUL_4556|M.ulcerans_Agy99 VHSHYWLSGQVGWLARDRWAVPLVHTAHT-LAAVKNAALANGDAPEPPLR
MAV_2755|M.avium_104 VHDNQSLG--TGLLSIAGSGLPVVATVHHPITRDRVLDLAAARWWRKPLV
MMAR_2925|M.marinum_M VHDNQSLG--TGLLRIAAMGLPVVATVHHPITRDRVLDLAAARWWRKPLV
MSMEG_6484|M.smegmatis_MC2_155 VHDNQSLG--TGLLKIEKLGLPVVATVHHPITRDKVVDVAAAKWWRKPLV
TH_0088|M.thermoresistible__bu VHDNQSLG--TGLLELADLGLPVVATVHHPITRDKVVEVAAAKWWRKPLV
Mflv_1099|M.gilvum_PYR-GCK VHDNQCLG--TGLLTIAESGMPVVATVHHPITRDKVLEVAAAKWWRKPLV
Mvan_5718|M.vanbaalenii_PYR-1 VHDNQCLG--TGLLTIADSGMPVVATVHHPITRDRVLEVAAAKWWRKPLV
MAB_1351c|M.abscessus_ATCC_199 VHSHTWYTGMAGHLAKLLHGIPHVVTAHSLEPRRPWKAEQLGGGYR----
**.: .* * .:* * *.* . . .
Mb0496|M.bovis_AF2122/97 ------TVGEQQVVDEADRLIVNTDDEARQVISLHG-ADPARIDVVHPGV
Rv0486|M.tuberculosis_H37Rv ------TVGEQQVVDEADRLIVNTDDEARQVISLHG-ADPARIDVVHPGV
MLBr_02443|M.leprae_Br4923 ------SVGEQQVVDEADRMIVNTDDEARQLISIHR-ADPAKIDVAHPGV
MUL_4556|M.ulcerans_Agy99 ------TVGEQQVVDEADRLIVNTDDEAKQLISIHH-ADPARIDVVHPGV
MAV_2755|M.avium_104 RRWYGFLAMQQEVARHIPDLLTVSSSSAADIISDFG-VSPEQLHVVPLGV
MMAR_2925|M.marinum_M HRWYGFSAMQKQVARQLPHLLTVSSSSAADIVTDFG-VSPDQLNVVPLGV
MSMEG_6484|M.smegmatis_MC2_155 RRWYGFAEMQKKVARQIPELLTVSSSSAADIAADFG-VTSDQLHIVPLGV
TH_0088|M.thermoresistible__bu RRWYGFAEMQKRVARRIPELITVSSSSAADISADFG-VSPGQLHVVPLGV
Mflv_1099|M.gilvum_PYR-GCK RRWYGFAEMQKDVARKIPELVTVSSTSAADIAEDFG-VTPEQLNVVPLGV
Mvan_5718|M.vanbaalenii_PYR-1 RRWYGFAEMQKQVARQIPELVTVSSTSAADIAEDFA-VDPNQLNVVPLGV
MAB_1351c|M.abscessus_ATCC_199 ----VSSWVEHTAIHAADAVIAVSGGMRDDIIATYPGLDPDRIHVVRNGI
:: . ::. :. :: . . ::.:. *:
Mb0496|M.bovis_AF2122/97 DLDVFRPGDRRAARA-------ALGLPVDERVVAFVGRIQPLKAPDIVLR
Rv0486|M.tuberculosis_H37Rv DLDVFRPGDRRAARA-------ALGLPVDERVVAFVGRIQPLKAPDIVLR
MLBr_02443|M.leprae_Br4923 DLDMFRPGDRRAARA-------ALGLPLDGNVVAFVGRIQPLKAPDIVLR
MUL_4556|M.ulcerans_Agy99 DLEVFRPGDRQQART-------ALGLRPEEKVVAFVGRIQPLKAPDIVLR
MAV_2755|M.avium_104 NTELFKPG---------------SGPRQPGRIIAIASADTPLKGVRTLLH
MMAR_2925|M.marinum_M NTELFEPG---------------VSPRKPGRVMAIASADTPLKGVSTLLH
MSMEG_6484|M.smegmatis_MC2_155 NTELFKP----------------AEQRVSGRIIAIASADVPLKGVSHLLH
TH_0088|M.thermoresistible__bu DTELFTP----------------SEHRVPGRIIAIASADVPLKGISHLLH
Mflv_1099|M.gilvum_PYR-GCK DTQLFQP----------------AEHRVRNRIIAIASADVPLKGVSHLLH
Mvan_5718|M.vanbaalenii_PYR-1 DTQLFQP----------------SEHRVRNRIIAIASADVPLKGVSHLLH
MAB_1351c|M.abscessus_ATCC_199 DTTSWYPVDPEEAFAQPGSVLAGLGVDLTRPIVAFVGRITRQKGLAHLVA
: : * ::*:.. *. ::
Mb0496|M.bovis_AF2122/97 AAAKLPGVRIIVAGGPSGSGLASPDGLVRLADELGIS--ARVTFLPPQSH
Rv0486|M.tuberculosis_H37Rv AAAKLPGVRIIVAGGPSGSGLASPDGLVRLADELGIS--ARVTFLPPQSH
MLBr_02443|M.leprae_Br4923 AAAKLPQVRIVVAGGPSGSGLASPDGLVRLADELGIT--ARVTFLPPQSR
MUL_4556|M.ulcerans_Agy99 AVAKLPGVRIIVAGGPSGSGLASPDGLAQLADELGIA--ERVTFLPPQSR
MAV_2755|M.avium_104 AVARLRTVRDLELRLVAKVEPNGP--THKLIAELGIS--DIVHITSGLSD
MMAR_2925|M.marinum_M AVAKLRVNREIELRLVAKVEPNGP--TEKLIAELGIS--DIVHISNGLSD
MSMEG_6484|M.smegmatis_MC2_155 AVARLRVERNLDLQLVSKLEPNGP--TEKLIAELGIS--DIVHISSGLSD
TH_0088|M.thermoresistible__bu AVARLRAKRNLDVQLVSKLEPNGP--TEKLIAELGIS--DIVQASSGLSD
Mflv_1099|M.gilvum_PYR-GCK AVARLRVERDLELQLVAKLEPNGP--TEKLIAELGIS--DIVHISSGLSD
Mvan_5718|M.vanbaalenii_PYR-1 AVARLRVERDLELQLVAKLEANGP--TEKLIAELGIS--DIVHISSGLSD
MAB_1351c|M.abscessus_ATCC_199 AAHRFSPEVQLILCAGEPDTPEIAEQITGAVTELAAARPGVHWIREFLPT
*. :: : . **. : .
Mb0496|M.bovis_AF2122/97 TDLATLFRAADLVAVPSYSESFGLVAVEAQACGTPVVAAAVGGLPVAVR-
Rv0486|M.tuberculosis_H37Rv TDLATLFRAADLVAVPSYSESFGLVAVEAQACGTPVVAAAVGGLPVAVR-
MLBr_02443|M.leprae_Br4923 TNLATVFQAADLVAVPSYSESFGLVAVEAQACGTPVVAAAVGGLPVAVR-
MUL_4556|M.ulcerans_Agy99 TDLARVFHAVDLVAIPSYSESFGLVAVEAQACGTRVVAAAVGGLPVAVR-
MAV_2755|M.avium_104 AELAALFASAEVACIPSLYEGFSLPAVEAMASGTPIVASRVGALPEVLGT
MMAR_2925|M.marinum_M AQLAALFASAEVACIPSLYEGFSLPAVEAMASGTPIVASRVGALPEVLGT
MSMEG_6484|M.smegmatis_MC2_155 QELAGLLASAEVACIPSLYEGFSLPAVEAMASGTPIVASRAGALPEVVGT
TH_0088|M.thermoresistible__bu SELARLLASAEVACIPSLYEGFSLPAVEAMASGTPIVASRAGALPEVLGP
Mflv_1099|M.gilvum_PYR-GCK SELAGLLASAEVACIPSLYEGFSLPAVEAMASGTPIVASRAGALPEVVGA
Mvan_5718|M.vanbaalenii_PYR-1 SELAALLASAEVACIPSLYEGFSLPAVEAMASGTPIVASRAGALPEVVGT
MAB_1351c|M.abscessus_ATCC_199 PKLREVLSAATVFVCPSIYEPLGIVNLEAMACGTAVVASRVGGIPEVVA-
.* :: :. : ** * :.: :** *.** :**: .*.:* .:
Mb0496|M.bovis_AF2122/97 DGITGTLVSGHE------VGQWADAIDHLLRLCAGPRGRVMSRAAARHAA
Rv0486|M.tuberculosis_H37Rv DGITGTLVSGHE------VGQWADAIDHLLRLCAGPRGRVMSRAAARHAA
MLBr_02443|M.leprae_Br4923 DGVTGTLVFGHN------VGHWADAVDQLLRLSAGPQARAISRAAVVHAA
MUL_4556|M.ulcerans_Agy99 DGVSGTLVSGHD------VDQWAAAIDGLLRSNAGAQGALMSRAAAEHAA
MAV_2755|M.avium_104 DGACAELVPPAD------VDALTRALGELLDSPEKRR-SLGRAGRTRAVD
MMAR_2925|M.marinum_M DGACAQLVPAAD------VGALTYALGELLDSPDKRR-SLGEAGRQRALG
MSMEG_6484|M.smegmatis_MC2_155 DGSCARLVRPAD------VDELTAVLGELLDSPAERR-RLGAAGRQRALD
TH_0088|M.thermoresistible__bu DGECARLVPPAD------VDALTRVLGELLDSPHQRS-RLGAAGRQRALD
Mflv_1099|M.gilvum_PYR-GCK DGQCARLVKPAD------VAELTSVLGELLDSPRELR-RLGDNGRRRALE
Mvan_5718|M.vanbaalenii_PYR-1 DGECARLVTPAD------VTELTAVLGELLDSPRELR-RLGDNGRRRALE
MAB_1351c|M.abscessus_ATCC_199 DGVTGTLVAYESSEAAAFEAGLADAVNALVADPARAT-AYGNAGRARCIE
** . ** . : .:. *: .
Mb0496|M.bovis_AF2122/97 TFSWENTTDALLASYRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRR
Rv0486|M.tuberculosis_H37Rv TFSWENTTDALLASYRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRR
MLBr_02443|M.leprae_Br4923 QFSWDNTTDALLASYRRAIGDFTATRQHR----VRDLVATRKPRRWISRR
MUL_4556|M.ulcerans_Agy99 TFSWENTTDALLASYRRAIGDFTAGRRRK----VRDPVAARKPRRWTARR
MAV_2755|M.avium_104 VFSWEAVAAQTVRVYERAIARNAAG---RAPC------------------
MMAR_2925|M.marinum_M VFSWEAVAAQTVRVYAAAIERTTTAPESNAQC------------------
MSMEG_6484|M.smegmatis_MC2_155 VFSWESVAAQTVAVYERAQNRMGVKAC-----------------------
TH_0088|M.thermoresistible__bu VFSWESVAAQTVSIYERAIC------------------------------
Mflv_1099|M.gilvum_PYR-GCK VFSWESVAAQTVAVYERACERVATC-------------------------
Mvan_5718|M.vanbaalenii_PYR-1 VFSWESVAAQTVAVYERACERVATC-------------------------
MAB_1351c|M.abscessus_ATCC_199 EFSWAHIAEMTLQIYQKVCS------------------------------
*** : : * .
Mb0496|M.bovis_AF2122/97 GVGA
Rv0486|M.tuberculosis_H37Rv GVGA
MLBr_02443|M.leprae_Br4923 GMGA
MUL_4556|M.ulcerans_Agy99 GVGA
MAV_2755|M.avium_104 ----
MMAR_2925|M.marinum_M ----
MSMEG_6484|M.smegmatis_MC2_155 ----
TH_0088|M.thermoresistible__bu ----
Mflv_1099|M.gilvum_PYR-GCK ----
Mvan_5718|M.vanbaalenii_PYR-1 ----
MAB_1351c|M.abscessus_ATCC_199 ----