For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLSVKVIGRAGKRITGESGPRPDSRAPPHPNYGAGMRVAMMTREYPPEVYGGAGVHVTELVTQLRRLCAV DVHCMGALRPGAAGVQVHQPDPRLQGANPALSTLSADLMMANAAAQASVVHSHTWYTGMAGHLAALLHDI PHVLTAHSLEPLRPWKAEQLGGGYRISTWVEHTAVLAAHAVIAVSSAMREDILRVYPALDPSAVHVIRNG VDTDVWYPAGPMQTGSMLAELGVDPARPLVVFVGRITRQKGLAHLVAAAHRFSPEAQVLLCAGAPDTPEI ADEIRDAVAALARGRTGVFWIREMLPIGELREILSAASVFVCPSVYEPLGIVNLEAMACAAPVVASDVGG IPEVVADGVTGSLVHYDPRDPAGYQAGLADAVNELIADREKAQRYGRAGRQRCVEEFSWAHVAEQTLEIY RKVCA*
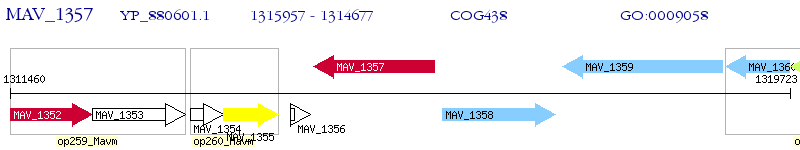
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1357 | - | - | 100% (426) | glycogen synthase |
| M. avium 104 | MAV_4082 | - | 1e-34 | 31.84% (424) | transferase |
| M. avium 104 | MAV_3879 | - | 2e-31 | 29.41% (425) | glycosyl transferase, group 1 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1244c | - | 0.0 | 78.46% (390) | putative glycosyl transferase |
| M. gilvum PYR-GCK | Mflv_2191 | - | 1e-163 | 73.25% (385) | glycogen synthase |
| M. tuberculosis H37Rv | Rv1212c | - | 0.0 | 78.72% (390) | putative glycosyl transferase |
| M. leprae Br4923 | MLBr_01715 | - | 3e-30 | 28.98% (421) | putative transferase |
| M. abscessus ATCC 19977 | MAB_1351c | - | 1e-167 | 73.03% (393) | putative glycosyltransferase |
| M. marinum M | MMAR_4226 | - | 1e-179 | 76.61% (389) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_5080 | - | 1e-171 | 75.13% (390) | glycogen synthase |
| M. thermoresistible (build 8) | TH_0550 | - | 1e-168 | 74.74% (388) | glycogen synthase |
| M. ulcerans Agy99 | MUL_1919 | - | 1e-32 | 30.07% (419) | transferase |
| M. vanbaalenii PYR-1 | Mvan_4504 | - | 1e-173 | 75.77% (388) | glycogen synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1244c|M.bovis_AF2122/97 ------------------------------------MRVAMLTREYPPEV
Rv1212c|M.tuberculosis_H37Rv ------------------------------------MRVAMLTREYPPEV
MAV_1357|M.avium_104 MRLSVKVIGRAGKRITGESGPRPDSRAPPHPNYGAGMRVAMMTREYPPEV
MMAR_4226|M.marinum_M ------------------------------------MRVAMLTREYPPEV
Mflv_2191|M.gilvum_PYR-GCK -----------------------------------------MTREYPPEV
Mvan_4504|M.vanbaalenii_PYR-1 ------------------------------------MRVAMMTREYPPEV
MSMEG_5080|M.smegmatis_MC2_155 ------------------------------------MRVAMMTREYPPEV
TH_0550|M.thermoresistible__bu ------------------------------------MRVAMMTREYPPEV
MAB_1351c|M.abscessus_ATCC_199 -----------------------------------MTRVAMMTREYPPEV
MLBr_01715|M.leprae_Br4923 ------------MFIQRHAVVRGIFGTFAPAEGTTLMKILIVSWEYPPVV
MUL_1919|M.ulcerans_Agy99 ------------------------------------MKILMVSWEYPPVV
:: **** *
Mb1244c|M.bovis_AF2122/97 YGGAGVHVTELVAYLRRLCAVDVHCMGAPRPGAF----------------
Rv1212c|M.tuberculosis_H37Rv YGGAGVHVTELVAYLRRLCAVDVHCMGAPRPGAF----------------
MAV_1357|M.avium_104 YGGAGVHVTELVTQLRRLCAVDVHCMGALRPGAAGVQ-------------
MMAR_4226|M.marinum_M YGGAGVHVTELVARLQRLCTVDVHCMGAPRPDAS----------------
Mflv_2191|M.gilvum_PYR-GCK YGGAGVHVTELVAQLRRLCEVDVHCMGAPRPDAM----------------
Mvan_4504|M.vanbaalenii_PYR-1 YGGAGVHVTELVAQLRHLCEVDVHCMGTPRPGVT----------------
MSMEG_5080|M.smegmatis_MC2_155 YGGAGVHVTELVAQLRKLCDVDVHCMGAPRDGAY----------------
TH_0550|M.thermoresistible__bu YGGAGVHVTELVAQLRHLCAVDVLCMGAPRVDAF----------------
MAB_1351c|M.abscessus_ATCC_199 YGGAGVHVTELVAQLRTLCDVDVHCMGAPRDNAF----------------
MLBr_01715|M.leprae_Br4923 IGGLGRHVHHLSTALAAAGHDVVVLSRRPSGTDPCTHPTSDEISEGVRVI
MUL_1919|M.ulcerans_Agy99 IGGLGRHVHHLATALVAAGHEVVVLSRRPSGTDPSTHPSSDLISEGVRVI
** * ** .* : * *
Mb1244c|M.bovis_AF2122/97 --AYRPDPRLGSANAALSTLSADLVMANAASAAT-----------VVHSH
Rv1212c|M.tuberculosis_H37Rv --AYRPDPRLGSANAALSTLSADLVMANAASAAT-----------VVHSH
MAV_1357|M.avium_104 --VHQPDPRLQGANPALSTLSADLMMANAAAQAS-----------VVHSH
MMAR_4226|M.marinum_M --AHQPDPRLANANPALATLSADLVIANAADAAS-----------VVHSH
Mflv_2191|M.gilvum_PYR-GCK --VAATDPALAGANPALTTLSADLNMVNAAAQAQ-----------VVHSH
Mvan_4504|M.vanbaalenii_PYR-1 --VAQPDAALVGANPALATLSADLNMVNSASAAT-----------VVHSH
MSMEG_5080|M.smegmatis_MC2_155 --VAHPDPTLRGANAALTMLSADLNMVNNAEAAT-----------VVHSH
TH_0550|M.thermoresistible__bu --AARPDPTLKGANAALSTLSADLHLVNAAGAAT-----------VVHSH
MAB_1351c|M.abscessus_ATCC_199 --VHRPDPALDGANPALTTLSADLVMAAAAAGST-----------VVHSH
MLBr_01715|M.leprae_Br4923 AAAQDPHEFTFSNDMMAWTLAMGHAMIRTGLSLTRHSSDLPWRPDVVHAH
MUL_1919|M.ulcerans_Agy99 AAAQDPHEFSFGSDMMAWTLAMGHAMVRAGLTLSGPGAEQPWQPDIVHAH
. .. . : *: . : . :**:*
Mb1244c|M.bovis_AF2122/97 TWYTALAGHLAAILYDIPHILTAHSLEPLRPWKKEQLGGGYQVSTWVEQT
Rv1212c|M.tuberculosis_H37Rv TWYTALAGHLAAILYDIPHVLTAHSLEPLRPWKKEQLGGGYQVSTWVEQT
MAV_1357|M.avium_104 TWYTGMAGHLAALLHDIPHVLTAHSLEPLRPWKAEQLGGGYRISTWVEHT
MMAR_4226|M.marinum_M TWYTGLAGHLAALLYGIPHVLTAHSLEPMRPWKTEQLGGGYQISSWVEKT
Mflv_2191|M.gilvum_PYR-GCK TWYAGMAGHLSALLYGVPHVLTAHSLEPMRPWKAEQLGGGYRISSWVEKT
Mvan_4504|M.vanbaalenii_PYR-1 TWYAGMAGHLAALLYGVPHVLTAHSLEPMRPWKAEQLGGGYRISSWVEKT
MSMEG_5080|M.smegmatis_MC2_155 TWYTGLAGHLASLLYGVPHVLTAHSLEPLRPWKAEQLGGGYQVSSWVERT
TH_0550|M.thermoresistible__bu TWYTGLAGHLAALLYGIPHVLTAHSLEPMRPWKAEQLGGGYRISSWVERT
MAB_1351c|M.abscessus_ATCC_199 TWYTGMAGHLAKLLHGIPHVVTAHSLEPRRPWKAEQLGGGYRVSSWVEHT
MLBr_01715|M.leprae_Br4923 DWLVAHPAITLAQFYDVPMVSTIHATEAGR-HSGWVSGALSRQVHAVESW
MUL_1919|M.ulcerans_Agy99 DWLVAHPAIALADYYDVPMVSTIHATEAGR-HSGWVSGDLSRQVHAVESW
* .. .. :.:* : * *: *. * . * : **
Mb1244c|M.bovis_AF2122/97 AVLAANAVIAVSSAMRNDMLRVYPSLDPNLVHVIRNGIDTETWYPAGP--
Rv1212c|M.tuberculosis_H37Rv AVLAANAVIAVSSAMRNDMLRVYPSLDPNLVHVIRNGIDTETWYPAGP--
MAV_1357|M.avium_104 AVLAAHAVIAVSSAMREDILRVYPALDPSAVHVIRNGVDTDVWYPAGP--
MMAR_4226|M.marinum_M AVLAADAVIAVSSGMRDDVLRLYPGLDPGMVHVVRNGIDTEVWFPAGP--
Mflv_2191|M.gilvum_PYR-GCK AVEAADAVIAVSAGMREDVLHAYPTLDPSRVHVVLNGIDTDKWRPVDA--
Mvan_4504|M.vanbaalenii_PYR-1 AVEAADAVIAVSSGMRADVLRTYPALDPERVHVVRNGIDTEVWYPAEP--
MSMEG_5080|M.smegmatis_MC2_155 AVEAADAVIAVSSGMRDDVLRTYPALDPDRVHVVRNGIDTTVWYPAEP--
TH_0550|M.thermoresistible__bu AVEAADAVIAVSSGMRADVLRTYPAVDPARVHVVRNGIDTDVWYPAPP--
MAB_1351c|M.abscessus_ATCC_199 AIHAADAVIAVSGGMRDDIIATYPGLDPDRIHVVRNGIDTTSWYPVDPEE
MLBr_01715|M.leprae_Br4923 LVRESDSLITCSASMCNEIIELFGPG-LAEITVIRNGIDPARWPFAAR--
MUL_1919|M.ulcerans_Agy99 LVRESDSLITCSGSMSDEITELFGPG-LAETAVIRNGIDVARWPFAVR--
: :.::*: *..* :: : *: **:* * .
Mb1244c|M.bovis_AF2122/97 --ARTGSVLAELGVDPNRPMAVFVGRITRQKGVVHLVTAAHRFRSDVQLV
Rv1212c|M.tuberculosis_H37Rv --ARTGSVLAELGVDPNRPMAVFVGRITRQKGVVHLVTAAHRFRSDVQLV
MAV_1357|M.avium_104 --MQTGSMLAELGVDPARPLVVFVGRITRQKGLAHLVAAAHRFSPEAQVL
MMAR_4226|M.marinum_M --VGTASVLAELGVDPNRPIVAFVGRITRQKGVPHLLAAAHQFSPDVQLV
Mflv_2191|M.gilvum_PYR-GCK --GGDESVLTELGVDLSRPIVAFVGRITRQKGVAHLVAAAHHFDPGIQLV
Mvan_4504|M.vanbaalenii_PYR-1 --DTDESVLAKLGVDPTRPVVAFVGRITRQKGVAHLVAAAHHFAPEVQLV
MSMEG_5080|M.smegmatis_MC2_155 --GPDESVLAELGVDLNRPIVAFVGRITRQKGVAHLVAAAHRFAPDVQLV
TH_0550|M.thermoresistible__bu --DPQESVLAELGVDPGRPIVAFVGRITRQKGVAHLLAAAHRFDPGIQLV
MAB_1351c|M.abscessus_ATCC_199 AFAQPGSVLAGLGVDLTRPIVAFVGRITRQKGLAHLVAAAHRFSPEVQLI
MLBr_01715|M.leprae_Br4923 ------------RARTGPAELLYVGRLEYEKGVHDVIAALPRIRRSYPGT
MUL_1919|M.ulcerans_Agy99 ------------KPRTGPPELLYVGRLEYEKGVHNAIAALPRIRRAHPGT
. :***: :**: . ::* ::
Mb1244c|M.bovis_AF2122/97 LCAGAADTPEVADEVRVAVAELARNRTGVFWIQDRLTIGQLREILSAATV
Rv1212c|M.tuberculosis_H37Rv LCAGAADTPEVADEVRVAVAELARNRTGVFWIQDRLTIGQLREILSAATV
MAV_1357|M.avium_104 LCAGAPDTPEIADEIRDAVAALARGRTGVFWIREMLPIGELREILSAASV
MMAR_4226|M.marinum_M LCAGAPDTPEIANEVQSAVAQLASTRSGVFWIRDILPVQKLREILSAATV
Mflv_2191|M.gilvum_PYR-GCK LCAGAPDTPEIAEEVASAVQRLAEARTGVFWVREMLPQPKIDEILSAATT
Mvan_4504|M.vanbaalenii_PYR-1 LCAGAPDTPEIAEEVASAVQALSQARTGVFWVREMLPTPKIREILSAATT
MSMEG_5080|M.smegmatis_MC2_155 LCAGAPDTPQIAEEVSSAVQQLAQARTGVFWVREMLPTHKIREILSAATV
TH_0550|M.thermoresistible__bu LCAGAPDTPEIAAEITAAVGELARRRTGVFWVRDMLPIGKIREILSAATV
MAB_1351c|M.abscessus_ATCC_199 LCAGEPDTPEIAEQITGAVTELAAARPGVHWIREFLPTPKLREVLSAATV
MLBr_01715|M.leprae_Br4923 TLTIAGEGTQQDWLVDQARKYKVIKAT---RFVGHLNHNELLAALQRADA
MUL_1919|M.ulcerans_Agy99 TLTIAGEGTQQDWLVEQARKHRVLKAT---KFLGHLEHAELLTLLHRADA
: : .: : * . . * :: * * .
Mb1244c|M.bovis_AF2122/97 FVCPSVYEPLGIVNLEAMACATAVVASDVGGIPEVVADGITGSLVHYDAD
Rv1212c|M.tuberculosis_H37Rv FVCPSVYEPLGIVNLEAMACATAVVASDVGGIPEVVADGITGSLVHYDAD
MAV_1357|M.avium_104 FVCPSVYEPLGIVNLEAMACAAPVVASDVGGIPEVVADGVTGSLVHYDPR
MMAR_4226|M.marinum_M FVCASIYEPLGIVNLEAMACATAVVASDVGGIPEVVADGITGTLVHYAAD
Mflv_2191|M.gilvum_PYR-GCK FVCPSVYEPLGIVNLEAMACATAVVASDVGGIPEVVADGQTGLLVHYDAK
Mvan_4504|M.vanbaalenii_PYR-1 FVCPSVYEPLGIVNLEAMACATAVVASDVGGIPEVVVDQRTGLLVHYDPD
MSMEG_5080|M.smegmatis_MC2_155 FVCPSVYEPLGIVNLEAMACATAVVASDVGGIPEVVADGRTGLLVHYDAN
TH_0550|M.thermoresistible__bu FVCPSVYEPLGIVNLEAMACGTAVVASDVGGIPEVVSDHRTGLLVHYDAG
MAB_1351c|M.abscessus_ATCC_199 FVCPSIYEPLGIVNLEAMACGTAVVASRVGGIPEVVADGVTGTLVAYESS
MLBr_01715|M.leprae_Br4923 AVLPSHYEPFGLVALEAAAAGTPLVTSNIGGLGEAVINGQTG--VSCPPR
MUL_1919|M.ulcerans_Agy99 AVFPSHYEPFGLVALEAAAAGTPLVTSNIGGLGEAVIDGVTG--VSCPPR
* .* ***:*:* *** *..:.:*:* :**: *.* : ** * .
Mb1244c|M.bovis_AF2122/97 DATGYQARLAEAVNALVADPATAERYGHAGRQRCIQEFSWAYIAEQTLDI
Rv1212c|M.tuberculosis_H37Rv DATGYQARLAEAVNALVADPATAERYGHAGRQRCIQEFSWAYIAEQTLDI
MAV_1357|M.avium_104 DPAGYQAGLADAVNELIADREKAQRYGRAGRQRCVEEFSWAHVAEQTLEI
MMAR_4226|M.marinum_M DPAGYQSRLAQAVNALVADPAKAERYGQAGRQRCIEEFSWTQIAEQTLDI
Mflv_2191|M.gilvum_PYR-GCK DTATFERQLAEAVNSLVADPQRAREYGVAGRQRCIDEFSWARIAEQTLQI
Mvan_4504|M.vanbaalenii_PYR-1 DTASFERGLAEAVNSLVADPQRARRYGVDGRQRCIDEFSWAHIAEQTLQI
MSMEG_5080|M.smegmatis_MC2_155 DTEAYEARLAEAVNSLVADPDRAREYGVAGRERCIEEFSWAHIAEQTLEI
TH_0550|M.thermoresistible__bu DTEMYERRLAEAVNTLVSEPDRARQYGQAGRDRCIREFSWAHIAEQTLQI
MAB_1351c|M.abscessus_ATCC_199 EAAAFEAGLADAVNALVADPARATAYGNAGRARCIEEFSWAHIAEMTLQI
MLBr_01715|M.leprae_Br4923 D----IAELAAMVCTVLEDPDAAQQRALAARERLTSDFDWQTVAQQTAQV
MUL_1919|M.ulcerans_Agy99 D----VAALAAAVCGVLDDPAAAQQRARAARERLNSNFDWQTVAEQTAQV
: ** * :: : * . .* * :*.* :*: * ::
Mb1244c|M.bovis_AF2122/97 YRKVCA-------------------
Rv1212c|M.tuberculosis_H37Rv YRKVCA-------------------
MAV_1357|M.avium_104 YRKVCA-------------------
MMAR_4226|M.marinum_M YRKVCR-------------------
Mflv_2191|M.gilvum_PYR-GCK YRSVTA-------------------
Mvan_4504|M.vanbaalenii_PYR-1 YRKVLN-------------------
MSMEG_5080|M.smegmatis_MC2_155 YRKVSA-------------------
TH_0550|M.thermoresistible__bu YRQVAG-------------------
MAB_1351c|M.abscessus_ATCC_199 YQKVCS-------------------
MLBr_01715|M.leprae_Br4923 YLAAKRRERQPQPRLPIVEHALPDR
MUL_1919|M.ulcerans_Agy99 YLAAKRGERQPLPRLPIVEHALPDR
* .