For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIALLSYRSKTHCGGQGVYVRHLSSGLAELGHDVEVFSGQPYPEGLDPRVTLTKVPSLDLYREPDPFRVP HPREIRDGIDALELLTMWSAGFPEPRTFTMRAARILAARRDEFDVVHDNQSLGTGLLKIEKLGLPVVATV HHPITRDKVVDVAAAKWWRKPLVRRWYGFAEMQKKVARQIPELLTVSSSSAADIAADFGVTSDQLHIVPL GVNTELFKPAEQRVSGRIIAIASADVPLKGVSHLLHAVARLRVERNLDLQLVSKLEPNGPTEKLIAELGI SDIVHISSGLSDQELAGLLASAEVACIPSLYEGFSLPAVEAMASGTPIVASRAGALPEVVGTDGSCARLV RPADVDELTAVLGELLDSPAERRRLGAAGRQRALDVFSWESVAAQTVAVYERAQNRMGVKAC*
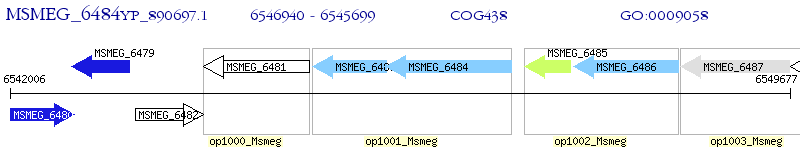
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6484 | - | - | 100% (413) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_0933 | - | 5e-21 | 28.40% (426) | hypothetical protein MSMEG_0933 |
| M. smegmatis MC2 155 | MSMEG_5080 | - | 1e-19 | 27.60% (337) | glycogen synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3058 | - | 2e-23 | 28.67% (429) | transferase |
| M. gilvum PYR-GCK | Mflv_1099 | - | 0.0 | 85.29% (408) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv3032 | - | 2e-23 | 28.67% (429) | transferase |
| M. leprae Br4923 | MLBr_01715 | - | 1e-22 | 25.85% (414) | putative transferase |
| M. abscessus ATCC 19977 | MAB_3366 | - | 5e-22 | 25.76% (427) | glycosyltransferase |
| M. marinum M | MMAR_2925 | - | 0.0 | 79.17% (408) | glycosyltransferase |
| M. avium 104 | MAV_2755 | - | 1e-176 | 79.04% (396) | glycosyl transferase |
| M. thermoresistible (build 8) | TH_0088 | - | 0.0 | 87.38% (404) | glycosyl transferase |
| M. ulcerans Agy99 | MUL_4556 | - | 2e-21 | 27.63% (427) | mannosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5718 | - | 0.0 | 84.31% (408) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6484|M.smegmatis_MC2_155 ------------------------MRIALLSYRS-KTHCGGQGVYVRHLS
TH_0088|M.thermoresistible__bu ------------------------MRIALLSYRS-KTHCGGQGVYVRHLS
Mflv_1099|M.gilvum_PYR-GCK ------------------------MRIALLSYRS-KTHCGGQGVYVRHLS
Mvan_5718|M.vanbaalenii_PYR-1 ------------------------MRIALLSYRS-KTHCGGQGVYVRHLS
MMAR_2925|M.marinum_M ------------------------MRIALLSYRS-KTHCGGQGVYVRHLS
MAV_2755|M.avium_104 ---------------------------------------------MRHLS
Mb3058|M.bovis_AF2122/97 ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
Rv3032|M.tuberculosis_H37Rv ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
MLBr_01715|M.leprae_Br4923 MFIQRHAVVRGIFGTFAPAEGTTLMKILIVSWEYPPVVIGGLGRHVHHLS
MAB_3366|M.abscessus_ATCC_1997 ------------------------MRILMVSWEYPPVVIGGLGRHVHHLS
MUL_4556|M.ulcerans_Agy99 --------MRAGAGAAGESCKDDGVRPDDVSRLTADAVVRRVALLAVHTS
* *
MSMEG_6484|M.smegmatis_MC2_155 SGLAELGHDVEVFSGQPYPEGLDPRVTLTKVPSLDLYREPDPFRVPHPRE
TH_0088|M.thermoresistible__bu RGLVELGHQVEVFSGQPYPEGLDPRVKLTKVPSLDLYREPDPFRIPRPSE
Mflv_1099|M.gilvum_PYR-GCK RGLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDPFRIPRPSE
Mvan_5718|M.vanbaalenii_PYR-1 RGLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDPFRIPRPSE
MMAR_2925|M.marinum_M RGLVDLGHDVEVFSGQPYPELLDPRVRLTEVPSLDLYREPDPFRVPHPSE
MAV_2755|M.avium_104 HGLVELGHDVEVFSGQPYPDLLDPRVRLTEVPSLDLYREPDPFRVPRPSE
Mb3058|M.bovis_AF2122/97 TALAAAGHDVVVLSRC--PSGTDP----STHPSSDEVTEG--VRVIAAAQ
Rv3032|M.tuberculosis_H37Rv TALAAAGHDVVVLSRC--PSGTDP----STHPSSDEVTEG--VRVIAAAQ
MLBr_01715|M.leprae_Br4923 TALAAAGHDVVVLSRR--PSGTDP----CTHPTSDEISEG--VRVIAAAQ
MAB_3366|M.abscessus_ATCC_1997 TELAAAGHDVVVLTRR--PSGTDP----SSHPTSDEISEG--VRVVAAAE
MUL_4556|M.ulcerans_Agy99 PLAQPGTGDAGGMNVYVLQSALHLAKRGIEVEIFTRATASADPPVVRVAP
:. :. . . :
MSMEG_6484|M.smegmatis_MC2_155 IRDGIDALELLTMWSAGFPEPRTFTMRAARILAARRDEFDVVHDNQSLGT
TH_0088|M.thermoresistible__bu IRDRIDLLELLTTWTAGFPEPKTFTMRMARILAERRDEFDVVHDNQSLGT
Mflv_1099|M.gilvum_PYR-GCK IRSSIDLEELLTTWTAGFPEPKTFSMRAARLLAARRGDFDVVHDNQCLGT
Mvan_5718|M.vanbaalenii_PYR-1 IKTSIDLEELLTTWTAGFPEPKTFSMRAARLLADRRDDFDVVHDNQCLGT
MMAR_2925|M.marinum_M IRDSIDLLELFTMWTAGFPEPRTFSLRVARLLADRVGDFDLVHDNQSLGT
MAV_2755|M.avium_104 IRDSIDLLELLTTWTAGFPEPRTFTLRAARLLAGRTADFDVVHDNQSLGT
Mb3058|M.bovis_AF2122/97 DPHEFTFGNDMMAWTLAMG---HAMIRAGLRLKKLGTDRSWRPDVVHAHD
Rv3032|M.tuberculosis_H37Rv DPHEFTFGNDMMAWTLAMG---HAMIRAGLRLKKLGTDRSWRPDVVHAHD
MLBr_01715|M.leprae_Br4923 DPHEFTFSNDMMAWTLAMG---HAMIRTGLSLTRHSSDLPWRPDVVHAHD
MAB_3366|M.abscessus_ATCC_1997 DPHDFDFGTDMMAWTLAMG---HAMVRAGLALGLKGSG-DWRPDVVHAHD
MUL_4556|M.ulcerans_Agy99 GVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAAHEPGHYDIVHSHY
. * *
MSMEG_6484|M.smegmatis_MC2_155 GLLKIEKLGLPVVATVHHPITRDKVVDVAAAKWWRKPLVRRWYGFAEMQK
TH_0088|M.thermoresistible__bu GLLELADLGLPVVATVHHPITRDKVVEVAAAKWWRKPLVRRWYGFAEMQK
Mflv_1099|M.gilvum_PYR-GCK GLLTIAESGMPVVATVHHPITRDKVLEVAAAKWWRKPLVRRWYGFAEMQK
Mvan_5718|M.vanbaalenii_PYR-1 GLLTIADSGMPVVATVHHPITRDRVLEVAAAKWWRKPLVRRWYGFAEMQK
MMAR_2925|M.marinum_M GLLRIAAMGLPVVATVHHPITRDRVLDLAAARWWRKPLVHRWYGFSAMQK
MAV_2755|M.avium_104 GLLSIAGSGLPVVATVHHPITRDRVLDLAAARWWRKPLVRRWYGFLAMQQ
Mb3058|M.bovis_AF2122/97 WLVAHPAIALAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVH---AVES
Rv3032|M.tuberculosis_H37Rv WLVAHPAIALAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVH---AVES
MLBr_01715|M.leprae_Br4923 WLVAHPAITLAQFYDVPMVSTIHATEAGRHSGWVSGALSRQVH---AVES
MAB_3366|M.abscessus_ATCC_1997 WLVAHPAIALAEHFDVPLVSTLHATEAGRHSGWVSGRISRQVH---SVEW
MUL_4556|M.ulcerans_Agy99 WLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALANGDAPEPPLR-TVGEQ
* * * . : . :
MSMEG_6484|M.smegmatis_MC2_155 KVARQIPELLTVSSSSAADIAADFGVTSDQLHIVPLGVNTELFKP-AEQR
TH_0088|M.thermoresistible__bu RVARRIPELITVSSSSAADISADFGVSPGQLHVVPLGVDTELFTP-SEHR
Mflv_1099|M.gilvum_PYR-GCK DVARKIPELVTVSSTSAADIAEDFGVTPEQLNVVPLGVDTQLFQP-AEHR
Mvan_5718|M.vanbaalenii_PYR-1 QVARQIPELVTVSSTSAADIAEDFAVDPNQLNVVPLGVDTQLFQP-SEHR
MMAR_2925|M.marinum_M QVARQLPHLLTVSSSSAADIVTDFGVSPDQLNVVPLGVNTELFEPGVSPR
MAV_2755|M.avium_104 EVARHIPDLLTVSSSSAADIISDFGVSPEQLHVVPLGVNTELFKPGSGPR
Mb3058|M.bovis_AF2122/97 WLVRESDSLITCSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRP
Rv3032|M.tuberculosis_H37Rv WLVRESDSLITCSASMNDEITELFGPGLAEITVIRNGIDAARWPFAARRP
MLBr_01715|M.leprae_Br4923 WLVRESDSLITCSASMCNEIIELFGPGLAEITVIRNGIDPARWPFAARRA
MAB_3366|M.abscessus_ATCC_1997 WLARESDSLITCSASMLDEVTQLFGPELPQVHVIRNGIDVTRWVFAPRPP
MUL_4556|M.ulcerans_Agy99 QVVDEADRLIVNTDDEAKQLISIHHADPARIDVVHPGVDLEVFRPGDRQQ
:. . *:. : :: . .: :: *:: :
MSMEG_6484|M.smegmatis_MC2_155 VSG--------RIIAIASADVPLKGVSHLLHAVARLR-VERNLDLQLVSK
TH_0088|M.thermoresistible__bu VPG--------RIIAIASADVPLKGISHLLHAVARLR-AKRNLDVQLVSK
Mflv_1099|M.gilvum_PYR-GCK VRN--------RIIAIASADVPLKGVSHLLHAVARLR-VERDLELQLVAK
Mvan_5718|M.vanbaalenii_PYR-1 VRN--------RIIAIASADVPLKGVSHLLHAVARLR-VERDLELQLVAK
MMAR_2925|M.marinum_M KPG--------RVMAIASADTPLKGVSTLLHAVAKLR-VNREIELRLVAK
MAV_2755|M.avium_104 QPG--------RIIAIASADTPLKGVRTLLHAVARLR-TVRDLELRLVAK
Mb3058|M.bovis_AF2122/97 RTG-P------AELLYVGRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGE
Rv3032|M.tuberculosis_H37Rv RTG-P------AELLYVGRLEYEKGVHDAIAALPRLRRTHPGTTLTIAGE
MLBr_01715|M.leprae_Br4923 RTG-P------AELLYVGRLEYEKGVHDVIAALPRIRRSYPGTTLTIAGE
MAB_3366|M.abscessus_ATCC_1997 ADGTP------PVLLFVGRLEYEKGIHDAIAALPKIRRAHPGTVLAVAGD
MUL_4556|M.ulcerans_Agy99 ARTALGLRPEEKVVAFVGRIQPLKAPDIVLRAVAKLPGVRIIVAGGPSGS
: .. *. : *:.:: ..
MSMEG_6484|M.smegmatis_MC2_155 LEPN-GPTEKLIAELGISDIVHISSGLSDQELAGLLASAEVACIPSLYEG
TH_0088|M.thermoresistible__bu LEPN-GPTEKLIAELGISDIVQASSGLSDSELARLLASAEVACIPSLYEG
Mflv_1099|M.gilvum_PYR-GCK LEPN-GPTEKLIAELGISDIVHISSGLSDSELAGLLASAEVACIPSLYEG
Mvan_5718|M.vanbaalenii_PYR-1 LEAN-GPTEKLIAELGISDIVHISSGLSDSELAALLASAEVACIPSLYEG
MMAR_2925|M.marinum_M VEPN-GPTEKLIAELGISDIVHISNGLSDAQLAALFASAEVACIPSLYEG
MAV_2755|M.avium_104 VEPN-GPTHKLIAELGISDIVHITSGLSDAELAALFASAEVACIPSLYEG
Mb3058|M.bovis_AF2122/97 GTQQ-DWLIDQARKHRVLRATRFVGHLDHTELLALLHRADAAVLPSHYEP
Rv3032|M.tuberculosis_H37Rv GTQQ-DWLIDQARKHRVLRATRFVGHLDHTELLALLHRADAAVLPSHYEP
MLBr_01715|M.leprae_Br4923 GTQQ-DWLVDQARKYKVIKATRFVGHLNHNELLAALQRADAAVLPSHYEP
MAB_3366|M.abscessus_ATCC_1997 GTQQ-DWLLEQARKYKVVKSVQFLGTLDHSELLHWLHHADAILLPSHYEP
MUL_4556|M.ulcerans_Agy99 GLASPDGLAQLADELGIAERVTFLPPQSRTDLARVFHAVDLVAIPSYSES
. . . : : . . :* : .: :** *
MSMEG_6484|M.smegmatis_MC2_155 FSLPAVEAMASGTPIVASRAGALPEVVGTDGSCARLVRPADVDELTAVLG
TH_0088|M.thermoresistible__bu FSLPAVEAMASGTPIVASRAGALPEVLGPDGECARLVPPADVDALTRVLG
Mflv_1099|M.gilvum_PYR-GCK FSLPAVEAMASGTPIVASRAGALPEVVGADGQCARLVKPADVAELTSVLG
Mvan_5718|M.vanbaalenii_PYR-1 FSLPAVEAMASGTPIVASRAGALPEVVGTDGECARLVTPADVTELTAVLG
MMAR_2925|M.marinum_M FSLPAVEAMASGTPIVASRVGALPEVLGTDGACAQLVPAADVGALTYALG
MAV_2755|M.avium_104 FSLPAVEAMASGTPIVASRVGALPEVLGTDGACAELVPPADVDALTRALG
Mb3058|M.bovis_AF2122/97 FGLVALEAAAAGTPLVTSNIGGLGEAV-INGQTGVSCAPRDVAGLAAAVR
Rv3032|M.tuberculosis_H37Rv FGLVALEAAAAGTPLVTSNIGGLGEAV-INGQTGVSCAPRDVAGLAAAVR
MLBr_01715|M.leprae_Br4923 FGLVALEAAAAGTPLVTSNIGGLGEAV-INGQTGVSCPPRDIAELAAMVC
MAB_3366|M.abscessus_ATCC_1997 FGIVALEAAAAGTPLVTSTVGGLGEAV-IDGETGMSFAPRDVAGIAEAVT
MUL_4556|M.ulcerans_Agy99 FGLVAVEAQACGTRVVAAAVGGLPVAV-RDGVSGTLVSGHDVDQWAAAID
*.: *:** *.** :*:: *.* .: :* . *: : :
MSMEG_6484|M.smegmatis_MC2_155 ELLDSPAERR-RLGAAGRQRALDVFSWESVAAQTVAVYERAQ------NR
TH_0088|M.thermoresistible__bu ELLDSPHQRS-RLGAAGRQRALDVFSWESVAAQTVSIYERAI------C-
Mflv_1099|M.gilvum_PYR-GCK ELLDSPRELR-RLGDNGRRRALEVFSWESVAAQTVAVYERAC------ER
Mvan_5718|M.vanbaalenii_PYR-1 ELLDSPRELR-RLGDNGRRRALEVFSWESVAAQTVAVYERAC------ER
MMAR_2925|M.marinum_M ELLDSPDKRR-SLGEAGRQRALGVFSWEAVAAQTVRVYAAAI------ER
MAV_2755|M.avium_104 ELLDSPEKRR-SLGRAGRTRAVDVFSWEAVAAQTVRVYERAI------AR
Mb3058|M.bovis_AF2122/97 SVLDDPAAAQ-RRARAARQRLTSDFDWQTVATATAQVYLAAK------RG
Rv3032|M.tuberculosis_H37Rv SVLDDPAAAQ-RRARAARQRLTSDFDWQTVATATAQVYLAAK------RG
MLBr_01715|M.leprae_Br4923 TVLEDPDAAQ-QRALAARERLTSDFDWQTVAQQTAQVYLAAK------RR
MAB_3366|M.abscessus_ATCC_1997 RTLDDPAAAA-ERAVAARARLTADFDWATVADETAQVYLAAK------RR
MUL_4556|M.ulcerans_Agy99 GLLRSNAGAQGALMSRAAAEHAATFSWENTTDALLASYRRAIGDFTAGRR
* . . . *.* .: * *
MSMEG_6484|M.smegmatis_MC2_155 MGVKAC-----------------
TH_0088|M.thermoresistible__bu -----------------------
Mflv_1099|M.gilvum_PYR-GCK VATC-------------------
Mvan_5718|M.vanbaalenii_PYR-1 VATC-------------------
MMAR_2925|M.marinum_M TTTAPESNAQC------------
MAV_2755|M.avium_104 NAAG---RAPC------------
Mb3058|M.bovis_AF2122/97 ERQPQPRLPIVEHALPDR-----
Rv3032|M.tuberculosis_H37Rv ERQPQPRLPIVEHALPDR-----
MLBr_01715|M.leprae_Br4923 ERQPQPRLPIVEHALPDR-----
MAB_3366|M.abscessus_ATCC_1997 EREPLGRSVIVERPLPDR-----
MUL_4556|M.ulcerans_Agy99 RKVRDPVAARKPRRWTARRGVGA