For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIALLSYRSKTHCGGQGVYVRHLSRGLVDLGHDVEVFSGQPYPELLDPRVRLTEVPSLDLYREPDPFRVP HPSEIRDSIDLLELFTMWTAGFPEPRTFSLRVARLLADRVGDFDLVHDNQSLGTGLLRIAAMGLPVVATV HHPITRDRVLDLAAARWWRKPLVHRWYGFSAMQKQVARQLPHLLTVSSSSAADIVTDFGVSPDQLNVVPL GVNTELFEPGVSPRKPGRVMAIASADTPLKGVSTLLHAVAKLRVNREIELRLVAKVEPNGPTEKLIAELG ISDIVHISNGLSDAQLAALFASAEVACIPSLYEGFSLPAVEAMASGTPIVASRVGALPEVLGTDGACAQL VPAADVGALTYALGELLDSPDKRRSLGEAGRQRALGVFSWEAVAAQTVRVYAAAIERTTTAPESNAQC*
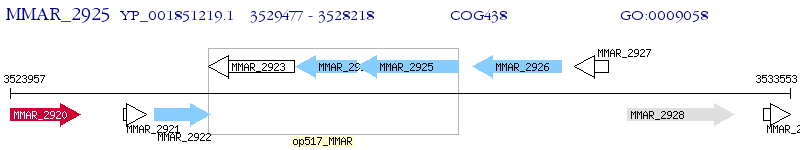
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2925 | - | - | 100% (419) | glycosyltransferase |
| M. marinum M | MMAR_0812 | - | 9e-23 | 28.44% (443) | mannosyltransferase |
| M. marinum M | MMAR_2092 | pimA | 1e-21 | 32.67% (300) | alpha-mannosyltransferase PimA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0496 | - | 1e-22 | 28.67% (443) | mannosyltransferase |
| M. gilvum PYR-GCK | Mflv_1099 | - | 0.0 | 78.83% (411) | glycosyl transferase, group 1 |
| M. tuberculosis H37Rv | Rv0486 | - | 1e-22 | 28.67% (443) | mannosyltransferase |
| M. leprae Br4923 | MLBr_00452 | - | 3e-23 | 32.58% (310) | putative glycosyltransferase |
| M. abscessus ATCC 19977 | MAB_4057c | - | 4e-21 | 27.87% (427) | glycosyl transferase |
| M. avium 104 | MAV_2755 | - | 0.0 | 86.08% (388) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_6484 | - | 0.0 | 79.17% (408) | glycosyl transferase |
| M. thermoresistible (build 8) | TH_0088 | - | 0.0 | 78.08% (406) | glycosyl transferase |
| M. ulcerans Agy99 | MUL_3252 | pimA | 1e-22 | 33.00% (300) | alpha-mannosyltransferase PimA |
| M. vanbaalenii PYR-1 | Mvan_5718 | - | 0.0 | 79.32% (411) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2925|M.marinum_M ---------------------------MRIALLSYRSKTHCGGQGVYVRH
MAV_2755|M.avium_104 -----------------------------------------------MRH
MSMEG_6484|M.smegmatis_MC2_155 ---------------------------MRIALLSYRSKTHCGGQGVYVRH
TH_0088|M.thermoresistible__bu ---------------------------MRIALLSYRSKTHCGGQGVYVRH
Mflv_1099|M.gilvum_PYR-GCK ---------------------------MRIALLSYRSKTHCGGQGVYVRH
Mvan_5718|M.vanbaalenii_PYR-1 ---------------------------MRIALLSYRSKTHCGGQGVYVRH
MLBr_00452|M.leprae_Br4923 --------------------------------------------------
MUL_3252|M.ulcerans_Agy99 --------------------------MAHVSATMVGRLVRCPPRAAAGAL
Mb0496|M.bovis_AF2122/97 MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
Rv0486|M.tuberculosis_H37Rv MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
MAB_4057c|M.abscessus_ATCC_199 -----------------MLSGDEFAR---------RAVAALK-PRRIAVL
MMAR_2925|M.marinum_M LSRGLVDLGHDVEVFSGQPYPELLDPRVRLTEVPSLDLYREPDPFRVPHP
MAV_2755|M.avium_104 LSHGLVELGHDVEVFSGQPYPDLLDPRVRLTEVPSLDLYREPDPFRVPRP
MSMEG_6484|M.smegmatis_MC2_155 LSSGLAELGHDVEVFSGQPYPEGLDPRVTLTKVPSLDLYREPDPFRVPHP
TH_0088|M.thermoresistible__bu LSRGLVELGHQVEVFSGQPYPEGLDPRVKLTKVPSLDLYREPDPFRIPRP
Mflv_1099|M.gilvum_PYR-GCK LSRGLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDPFRIPRP
Mvan_5718|M.vanbaalenii_PYR-1 LSRGLVELGHDVEVFSGQPYPEILDPRVRLTEVPSLDLYREPDPFRIPRP
MLBr_00452|M.leprae_Br4923 MRIGMICP-YSFDVPGGVQSHVLQLAEVMRARGQQVRVLAPASPDVSLPE
MUL_3252|M.ulcerans_Agy99 MRIGMVCP-YSFDVAGGVQSHVLQLAEVMRARGHEVSVLAPASPHAALPD
Mb0496|M.bovis_AF2122/97 AVHTSPLAQPGTGDAGGMNVYMLQSALHLARRGIEVEIFTRATASADPPV
Rv0486|M.tuberculosis_H37Rv AVHTSPLAQPGTGDAGGMNVYMLQSALHLARRGIEVEIFTRATASADPPV
MAB_4057c|M.abscessus_ATCC_199 SVHTSPLAQPGTGDAGGMNVYVLQTALQLARRGVAVDIFTRATSSSDEPV
.* . . : : . .
MMAR_2925|M.marinum_M SEIRDSIDLLELFTMWTAGFPEP---------RTFSLRVARLLADRVGDF
MAV_2755|M.avium_104 SEIRDSIDLLELLTTWTAGFPEP---------RTFTLRAARLLAGRTADF
MSMEG_6484|M.smegmatis_MC2_155 REIRDGIDALELLTMWSAGFPEP---------RTFTMRAARILAARRDEF
TH_0088|M.thermoresistible__bu SEIRDRIDLLELLTTWTAGFPEP---------KTFTMRMARILAERRDEF
Mflv_1099|M.gilvum_PYR-GCK SEIRSSIDLEELLTTWTAGFPEP---------KTFSMRAARLLAARRGDF
Mvan_5718|M.vanbaalenii_PYR-1 SEIKTSIDLEELLTTWTAGFPEP---------KTFSMRAARLLADRRDDF
MLBr_00452|M.leprae_Br4923 YVVSAGRAIPIPYNGSVARLQFS---------PAVHSRVRRWLVDGDFDV
MUL_3252|M.ulcerans_Agy99 YVVSGGKAVPIPYNGSVARLRFG---------PATHRKVKKWLSEGDFDV
Mb0496|M.bovis_AF2122/97 VRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPGYYDI
Rv0486|M.tuberculosis_H37Rv VRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPGYYDI
MAB_4057c|M.abscessus_ATCC_199 VSVTDGVTVRNIVAGPFEGLDKYDLPTQLCAFTAGVLRAEAVHEPGYYNL
: : : : :.
MMAR_2925|M.marinum_M DLVHDNQSLGTGLLRIAAMGLPVVATVHHPITRDRVLDLAAARWWRKPLV
MAV_2755|M.avium_104 DVVHDNQSLGTGLLSIAGSGLPVVATVHHPITRDRVLDLAAARWWRKPLV
MSMEG_6484|M.smegmatis_MC2_155 DVVHDNQSLGTGLLKIEKLGLPVVATVHHPITRDKVVDVAAAKWWRKPLV
TH_0088|M.thermoresistible__bu DVVHDNQSLGTGLLELADLGLPVVATVHHPITRDKVVEVAAAKWWRKPLV
Mflv_1099|M.gilvum_PYR-GCK DVVHDNQCLGTGLLTIAESGMPVVATVHHPITRDKVLEVAAAKWWRKPLV
Mvan_5718|M.vanbaalenii_PYR-1 DVVHDNQCLGTGLLTIADSGMPVVATVHHPITRDRVLEVAAAKWWRKPLV
MLBr_00452|M.leprae_Br4923 LHLHEPNAPSLSMWALRVAEGPIVATFHTSTTKSLTLSVFQG------VL
MUL_3252|M.ulcerans_Agy99 LHLHEPNAPSLSMLALNIAEGPIVATFHTSTTKSLTLTVFQS------IL
Mb0496|M.bovis_AF2122/97 VHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALADGDG--PEPPL
Rv0486|M.tuberculosis_H37Rv VHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALADGDG--PEPPL
MAB_4057c|M.abscessus_ATCC_199 VHSHYWLSGQAGWLARDRWGVPLVHTAHTLAAVKNQTLAEGDR--PEPAL
* . . *:* * * : . :
MMAR_2925|M.marinum_M HRWYGFSAMQKQVARQLPHLLTVSSSSAADIVTDFGVSPDQLNVVPLGVN
MAV_2755|M.avium_104 RRWYGFLAMQQEVARHIPDLLTVSSSSAADIISDFGVSPEQLHVVPLGVN
MSMEG_6484|M.smegmatis_MC2_155 RRWYGFAEMQKKVARQIPELLTVSSSSAADIAADFGVTSDQLHIVPLGVN
TH_0088|M.thermoresistible__bu RRWYGFAEMQKRVARRIPELITVSSSSAADISADFGVSPGQLHVVPLGVD
Mflv_1099|M.gilvum_PYR-GCK RRWYGFAEMQKDVARKIPELVTVSSTSAADIAEDFGVTPEQLNVVPLGVD
Mvan_5718|M.vanbaalenii_PYR-1 RRWYGFAEMQKQVARQIPELVTVSSTSAADIAEDFAVDPNQLNVVPLGVD
MLBr_00452|M.leprae_Br4923 RPWH------EKIIGRIAVSDLARRWQMEALGSDAVEIPNGVNVDSLSSA
MUL_3252|M.ulcerans_Agy99 RPMH------EKIVGRIAVSDLARRWQMEALGSDAVEIPNGVDVGSFASS
Mb0496|M.bovis_AF2122/97 RTVG-----EQQVVDEADRLIVNTDDEARQVISLHGADPARIDVVHPGVD
Rv0486|M.tuberculosis_H37Rv RTVG-----EQQVVDEADRLIVNTDDEARQVISLHGADPARIDVVHPGVD
MAB_4057c|M.abscessus_ATCC_199 RGVG-----EQQVVDEADRLIVNTKDEADQLVSIHNADPARIDIVHPGVD
: : : . . : . :.: .
MMAR_2925|M.marinum_M TELFEPG--------VSPRKPGRVMAIASADTPLKGVSTLLHAVAKLRVN
MAV_2755|M.avium_104 TELFKPG--------SGPRQPGRIIAIASADTPLKGVRTLLHAVARLRTV
MSMEG_6484|M.smegmatis_MC2_155 TELFKP---------AEQRVSGRIIAIASADVPLKGVSHLLHAVARLRVE
TH_0088|M.thermoresistible__bu TELFTP---------SEHRVPGRIIAIASADVPLKGISHLLHAVARLRAK
Mflv_1099|M.gilvum_PYR-GCK TQLFQP---------AEHRVRNRIIAIASADVPLKGVSHLLHAVARLRVE
Mvan_5718|M.vanbaalenii_PYR-1 TQLFQP---------SEHRVRNRIIAIASADVPLKGVSHLLHAVARLRVE
MLBr_00452|M.leprae_Br4923 PQLAGY-----------PRLGKTVLFLGRYDEPRKGMSVLLDALPGV-ME
MUL_3252|M.ulcerans_Agy99 SCLDGY-----------PRPGKTVLFLGRYDEPRKGMSVLLEALPKL-VE
Mb0496|M.bovis_AF2122/97 LDVFRPGDRRAARAALGLPVDERVVAFVGRIQPLKAPDIVLRAAAKLPGV
Rv0486|M.tuberculosis_H37Rv LDVFRPGDRRAARAALGLPVDERVVAFVGRIQPLKAPDIVLRAAAKLPGV
MAB_4057c|M.abscessus_ATCC_199 LDVFTPGDKAAARAEFGLRADEQVVAFVGRIQPLKAPDLLVRAAERLPGV
: :: : * *. :: * :
MMAR_2925|M.marinum_M REIELRLVAKVEPNGPT--EKLIAELG-ISDIVHISNGLSDAQLAALFAS
MAV_2755|M.avium_104 RDLELRLVAKVEPNGPT--HKLIAELG-ISDIVHITSGLSDAELAALFAS
MSMEG_6484|M.smegmatis_MC2_155 RNLDLQLVSKLEPNGPT--EKLIAELG-ISDIVHISSGLSDQELAGLLAS
TH_0088|M.thermoresistible__bu RNLDVQLVSKLEPNGPT--EKLIAELG-ISDIVQASSGLSDSELARLLAS
Mflv_1099|M.gilvum_PYR-GCK RDLELQLVAKLEPNGPT--EKLIAELG-ISDIVHISSGLSDSELAGLLAS
Mvan_5718|M.vanbaalenii_PYR-1 RDLELQLVAKLEANGPT--EKLIAELG-ISDIVHISSGLSDSELAALLAS
MLBr_00452|M.leprae_Br4923 CFDDVQLLIVGR--GDE--EQLRSQAGGLVEHIRFLGQVDDAGKAAAMRS
MUL_3252|M.ulcerans_Agy99 RFIDVQLLIVGR--GDE--DELRDEAGELAEHMRFLGQVDDAGKASAMRS
Mb0496|M.bovis_AF2122/97 RIIVAGGPSGSGLASPDGLVRLADELG-ISARVTFLPPQSHTDLATLFRA
Rv0486|M.tuberculosis_H37Rv RIIVAGGPSGSGLASPDGLVRLADELG-ISARVTFLPPQSHTDLATLFRA
MAB_4057c|M.abscessus_ATCC_199 RVLIVGGPSGSGLDEPTALQDLAVDLG-IADRVTFLPPQTRERLAQVYRA
* : * : : * :
MMAR_2925|M.marinum_M AEVACIPSLY-EGFSLPAVEAMASGTPIVASRVGALPEVLGTDGACAQLV
MAV_2755|M.avium_104 AEVACIPSLY-EGFSLPAVEAMASGTPIVASRVGALPEVLGTDGACAELV
MSMEG_6484|M.smegmatis_MC2_155 AEVACIPSLY-EGFSLPAVEAMASGTPIVASRAGALPEVVGTDGSCARLV
TH_0088|M.thermoresistible__bu AEVACIPSLY-EGFSLPAVEAMASGTPIVASRAGALPEVLGPDGECARLV
Mflv_1099|M.gilvum_PYR-GCK AEVACIPSLY-EGFSLPAVEAMASGTPIVASRAGALPEVVGADGQCARLV
Mvan_5718|M.vanbaalenii_PYR-1 AEVACIPSLY-EGFSLPAVEAMASGTPIVASRAGALPEVVGTDGECARLV
MLBr_00452|M.leprae_Br4923 ADVYCAPNIGGESFGIVLVEAMAAGTPVVASDLDAFRRVLR-DGEVGHLV
MUL_3252|M.ulcerans_Agy99 ADVYCAPNTGGESFGIVLVEAMAAGTPVVASDLDAFRRVLR-DGEIGRLV
Mb0496|M.bovis_AF2122/97 ADLVAVPSYS-ESFGLVAVEAQACGTPVVAAAVGGLPVAVR-DGITGTLV
Rv0486|M.tuberculosis_H37Rv ADLVAVPSYS-ESFGLVAVEAQACGTPVVAAAVGGLPVAVR-DGITGTLV
MAB_4057c|M.abscessus_ATCC_199 ADIVAVPSYS-ESFGLVAIEAQACGTPVVAAAVGGLPVAVA-DQRTGLLV
*:: . *. *.*.: :** *.***:**: ..: .: * . **
MMAR_2925|M.marinum_M PAADVGALTYALGELLDS-PDKRRSLGEAGRQRALGVFSWEAVAAQTVRV
MAV_2755|M.avium_104 PPADVDALTRALGELLDS-PEKRRSLGRAGRTRAVDVFSWEAVAAQTVRV
MSMEG_6484|M.smegmatis_MC2_155 RPADVDELTAVLGELLDS-PAERRRLGAAGRQRALDVFSWESVAAQTVAV
TH_0088|M.thermoresistible__bu PPADVDALTRVLGELLDS-PHQRSRLGAAGRQRALDVFSWESVAAQTVSI
Mflv_1099|M.gilvum_PYR-GCK KPADVAELTSVLGELLDS-PRELRRLGDNGRRRALEVFSWESVAAQTVAV
Mvan_5718|M.vanbaalenii_PYR-1 TPADVTELTAVLGELLDS-PRELRRLGDNGRRRALEVFSWESVAAQTVAV
MLBr_00452|M.leprae_Br4923 PAGDSAALADALVALLRN-DVLRERYVAAG-AEAVRRYDWSVVASQIMRV
MUL_3252|M.ulcerans_Agy99 PVDDSDALAAALIEVLDS-QELRERYVAAG-TEAVRRYDWSVVASQIMRV
Mb0496|M.bovis_AF2122/97 SGHEVGQWADAIDHLLRLCAGPRGRVMSRAAARHAATFSWENTTDALLAS
Rv0486|M.tuberculosis_H37Rv SGHEVGQWADAIDHLLRLCAGPRGRVMSRAAARHAATFSWENTTDALLAS
MAB_4057c|M.abscessus_ATCC_199 PTHRTEDWADAIGELLVR----KGAGFSRAAVEHAAGFSWSSTADSLLSS
: .: :* . . :.*. .: :
MMAR_2925|M.marinum_M YAAAIERTTTAPESNAQC----------------------
MAV_2755|M.avium_104 YERAIARNAAG---RAPC----------------------
MSMEG_6484|M.smegmatis_MC2_155 YERAQNRMGVKAC---------------------------
TH_0088|M.thermoresistible__bu YERAIC----------------------------------
Mflv_1099|M.gilvum_PYR-GCK YERACERVATC-----------------------------
Mvan_5718|M.vanbaalenii_PYR-1 YERACERVATC-----------------------------
MLBr_00452|M.leprae_Br4923 YETVATSGSKVQVAS-------------------------
MUL_3252|M.ulcerans_Agy99 YETVAGAGVKVQVAS-------------------------
Mb0496|M.bovis_AF2122/97 YRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRRGVGA
Rv0486|M.tuberculosis_H37Rv YRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRRGVGA
MAB_4057c|M.abscessus_ATCC_199 YGRAIADYRAP-QRPSTQWAS--RARFRPRRLSGLRR---
* .