For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGNPVAAADPCSDVSVVFARGTHQEPGLGNIGQAFVDSLTSQLGGRSVDVYAVNYPANDDYHNSANAGAD DASAHVQDTVAACPNSRIVLGGYSQGSTVIDLATNAMPPSVADHVAAVALFGEPSSGFSTMLWGGQPLPT INPLYGGKTISLCAPDDPICSGGGNIMAHVSYIDAGMTAHAATFAANHLNQGSPRT
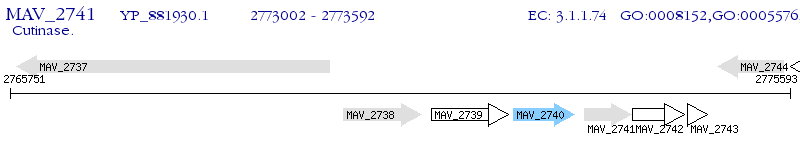
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2741 | - | - | 100% (196) | serine esterase, cutinase family protein |
| M. avium 104 | MAV_1682 | - | 3e-53 | 51.18% (211) | serine esterase, cutinase family protein |
| M. avium 104 | MAV_2961 | - | 3e-50 | 51.01% (198) | cutinase Cut3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2006c | cfp21 | 4e-86 | 77.25% (189) | cutinase precursor CFP21 |
| M. gilvum PYR-GCK | Mflv_3552 | - | 7e-46 | 48.73% (197) | cutinase |
| M. tuberculosis H37Rv | Rv1984c | cfp21 | 4e-86 | 77.25% (189) | cutinase precursor CFP21 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3272c | - | 6e-41 | 46.84% (190) | cutinase Cut1 |
| M. marinum M | MMAR_2934 | cfp21 | 6e-92 | 83.68% (190) | cutinase Cfp21 |
| M. smegmatis MC2 155 | MSMEG_2474 | - | 2e-43 | 45.41% (207) | cutinase Cut3 |
| M. thermoresistible (build 8) | TH_0238 | - | 1e-45 | 47.34% (207) | serine esterase, cutinase family protein |
| M. ulcerans Agy99 | MUL_1597 | cut1 | 9e-46 | 46.89% (209) | cutinase Cut1 |
| M. vanbaalenii PYR-1 | Mvan_3105 | - | 1e-53 | 53.50% (200) | cutinase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2741|M.avium_104 ----------------------MG---------------NPVAAADPCSD
MMAR_2934|M.marinum_M -------------MIARSFARILGAAVVA-LGVLSAPVFSPSATADPCSD
Mb2006c|M.bovis_AF2122/97 -------------MTPRSLVRIVGVVVATTLALVSAPAGGRAAHADPCSD
Rv1984c|M.tuberculosis_H37Rv -------------MTPRSLVRIVGVVVATTLALVSAPAGGRAAHADPCSD
Mflv_3552|M.gilvum_PYR-GCK --------------------MVTGWALSLSI------ASAPVAVAASCPD
Mvan_3105|M.vanbaalenii_PYR-1 -------------MQVRRVLGIVGAAVAVSAGLWPIGVGAPAASADPCPD
TH_0238|M.thermoresistible__bu -------LSRIGSAVGAALTTASLITGTVITSTVVAGTAVPAAAAQPCPD
MUL_1597|M.ulcerans_Agy99 ---------------------------------MFSAVAIPSASAEPCPD
MAB_3272c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_2474|M.smegmatis_MC2_155 MNNDHATTRPSIVRTAFVGVTLAALGLVMGVVGLQAAGTASAAPSSPCAD
MAV_2741|M.avium_104 VSVVFARGTHQEPGLGNIGQAFVDSLTSQLGGRSVDVYAVNYPANDDYHN
MMAR_2934|M.marinum_M VAVVFARGTHQEPGLGNIGQAFVDSLTSQVGGRSVDVYAVNYPANDDYHT
Mb2006c|M.bovis_AF2122/97 IAVVFARGTHQASGLGDVGEAFVDSLTSQVGGRSIGVYAVNYPASDDYRA
Rv1984c|M.tuberculosis_H37Rv IAVVFARGTHQASGLGDVGEAFVDSLTSQVGGRSIGVYAVNYPASDDYRA
Mflv_3552|M.gilvum_PYR-GCK VEVVFARGTADKPGVGFTGRQFVTALKKRLPGKTVSVYAVNYPASWNFSK
Mvan_3105|M.vanbaalenii_PYR-1 IEVIFARGTGAEPGLGWVGDAFVNALRPKVGGRSVGTYAVNYPAGFDFDN
TH_0238|M.thermoresistible__bu VQVVFARGTGEPPGVGPTGQAFIDALRPRVGDRSLDVYAVNYPAIDVWNT
MUL_1597|M.ulcerans_Agy99 VEVVFARGTGEPPGLGPTGQAFVNSLRSHIGGRSLDVYPVNYPASDNWDT
MAB_3272c|M.abscessus_ATCC_199 ---MFARGTFEPPGVGGTGESFVNALRARTKGKSVEVYPVEYPASLAFAT
MSMEG_2474|M.smegmatis_MC2_155 VEVVFARGTFEAPGVGDTGQAFVNALGAQLPGKNIAVYAVNYPASLDFND
:***** .*:* * *: :* : .:.: .*.*:*** :
MAV_2741|M.avium_104 SANAGADDASAHVQDTVAACPNSRIVLGGYSQGSTVIDLATN--------
MMAR_2934|M.marinum_M SASLGSDDASAHIQGTVANCPNTKIVLGGYSQGATVIDLAST--------
Mb2006c|M.bovis_AF2122/97 SASNGSDDASAHIQRTVASCPNTRIVLGGYSQGATVIDLSTS--------
Rv1984c|M.tuberculosis_H37Rv SASNGSDDASAHIQRTVASCPNTRIVLGGYSQGATVIDLSTS--------
Mflv_3552|M.gilvum_PYR-GCK STSEGAVDANKHVQYVAAVCPRTKIVLGGMSQGAGVIDLITVGNRII---
Mvan_3105|M.vanbaalenii_PYR-1 SAPLGAADASGRVQWMAGNCPDTKLVLGGMSQGAGVIDLITVDPRPL---
TH_0238|M.thermoresistible__bu GID-GIRDAGAHVVRMAEQCPDTRLVLGGFSQGAAVMGFVTSAEVPAGVD
MUL_1597|M.ulcerans_Agy99 GLD-GIRDAGTHVVSMAGQCPQTKMVLGGYSQGTAVMSFVTSAAVPAGVD
MAB_3272c|M.abscessus_ATCC_199 AAD-GVIDASNRVKDVAARCPNTKIVMGGFSQGAAVVAYTTADAVPPGFE
MSMEG_2474|M.smegmatis_MC2_155 AAL-GVADAAREVQAVAAGCPDTQIVLGGYSQGAAVAGYTTSGAVPAGYA
. * ** .: . ** :::*:** ***: * :
MAV_2741|M.avium_104 -------AMPPSVADHVAAVALFGEPSSGFSTMLWGGQPLPTINPLYGGK
MMAR_2934|M.marinum_M -------AMPSTVADHVAAVALFGEPSSGFSSMLWGGQPLPTINPLYGAK
Mb2006c|M.bovis_AF2122/97 -------AMPPAVADHVAAVALFGEPSSGFSSMLWGGGSLPTIGPLYSSK
Rv1984c|M.tuberculosis_H37Rv -------AMPPAVADHVAAVALFGEPSSGFSSMLWGGGSLPTIGPLYSSK
Mflv_3552|M.gilvum_PYR-GCK WFF-KPAPLPDKMVNHVAAIAVFGNPSRDIS----SLGPLTKISPLYGSK
Mvan_3105|M.vanbaalenii_PYR-1 GRF-TPTPMPPNVADHVAAVVVFGNPLRDIR----GGGPLPQMSGTYGSK
TH_0238|M.thermoresistible__bu PAT-VPKPLDPDIAEHVAAVVLFGMP--NARAMNFLGTPPVVIGPLYEDK
MUL_1597|M.ulcerans_Agy99 PAT-VPKPLQPDVAEHVAAVVLFGMP--NVRAMNFLGEPAVEIGPAYQAK
MAB_3272c|M.abscessus_ATCC_199 LPEGITGPMPAEVADHVAAVALFGKPSSGFLQRIDNTAPPINIGHLYVAK
MSMEG_2474|M.smegmatis_MC2_155 LPAGITGPMPPAIAPHVAAVVLFGTPSPFFLGLVDHSAPPLVIGPAYAAK
.: :. ****:.:** * . :. * *
MAV_2741|M.avium_104 TISLCAPDDPICS-GGGNIMAHVSYIDAG-MTAHAATFAANH-LNQGSPR
MMAR_2934|M.marinum_M TINLCAADDPICT-GGGNIMAHVSYIQAG-MTDQAASFAAGK-LN-----
Mb2006c|M.bovis_AF2122/97 TINLCAPDDPICT-GGGNIMAHVSYVQSG-MTSQAATFAANR-LDHAG--
Rv1984c|M.tuberculosis_H37Rv TINLCAPDDPICT-GGGNIMAHVSYVQSG-MTSQAATFAANR-LDHAG--
Mflv_3552|M.gilvum_PYR-GCK AIDLCATGDPYCS-SGNNFFAHFAYPWNG-MVNEAATFAAGRVLGTQN--
Mvan_3105|M.vanbaalenii_PYR-1 SIDLCAFDDPFCS-PGFNLPAHFAYVDNG-MVEEAANFAAGR-LG-----
TH_0238|M.thermoresistible__bu TVKLCAVDDPVCS-NGMNFAAHNTYATNAAMVDQGAAFAADR-LGVASDK
MUL_1597|M.ulcerans_Agy99 TIKVCVPEDPVCS-DGLNFAAHNAYADDGAVVDQGVAFAASR-LGGDPGG
MAB_3272c|M.abscessus_ATCC_199 TMDMCVPEDPICSPDGSDNAAHGSYGDNG-MTSQAADFAARQINS-----
MSMEG_2474|M.smegmatis_MC2_155 TLQLCNTGDPVCYPGGLDRAAHSAYKSNG-MADQAAQFVVDHLGATSTAT
::.:* ** * * : ** :* . :. ... *.. :
MAV_2741|M.avium_104 T------------
MMAR_2934|M.marinum_M -------------
Mb2006c|M.bovis_AF2122/97 -------------
Rv1984c|M.tuberculosis_H37Rv -------------
Mflv_3552|M.gilvum_PYR-GCK -------------
Mvan_3105|M.vanbaalenii_PYR-1 -------------
TH_0238|M.thermoresistible__bu PLPTPSSGQNFGN
MUL_1597|M.ulcerans_Agy99 PVPAPSSG--FGN
MAB_3272c|M.abscessus_ATCC_199 PTDTVSAAG----
MSMEG_2474|M.smegmatis_MC2_155 PAQVVSATTHP--