For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RIIVTGGNSGVGKATAHAMAAAGHEVVIACRSLTKAHQAAASMPGDVTVRELDLADLTSVRKFADTVDYV DVLVNNAGVLGLPLTRTADGFEAHMGINHLGHFALTCLLGDRIRDRVVLVASTNYNTARMHFDDLNYHHR RYNPWSAYGESKLANLLFVRELVARGKTAYASDPGMTDTGITRNGSGVLQWAGRVLSPHIAQSPADGARS EELSRPVDWGVSGDLRFG*
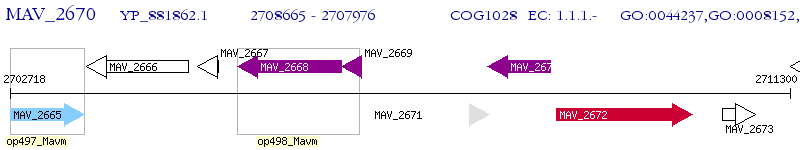
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2670 | - | - | 100% (229) | retinol dehydrogenase 12 |
| M. avium 104 | MAV_1968 | - | e-102 | 85.31% (211) | retinol dehydrogenase 12 |
| M. avium 104 | MAV_1153 | - | 3e-46 | 47.34% (207) | retinol dehydrogenase 13 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0447c | - | 2e-38 | 45.37% (227) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1940 | - | 8e-47 | 53.23% (186) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0439c | - | 2e-38 | 45.37% (227) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 6e-36 | 41.63% (245) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1144c | - | 2e-42 | 43.93% (214) | short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0757 | - | 9e-37 | 41.53% (248) | dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_5430 | - | 1e-47 | 48.15% (216) | retinol dehydrogenase 13 |
| M. thermoresistible (build 8) | TH_3199 | - | 2e-41 | 46.36% (220) | PUTATIVE retinol dehydrogenase 13 |
| M. ulcerans Agy99 | MUL_1391 | - | 2e-36 | 42.98% (228) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4786 | - | 3e-46 | 52.69% (186) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5430|M.smegmatis_MC2_155 -------MTQWTSADLPSFRGRTVVITGANSGLGLVTARELARVGAKVIV
TH_3199|M.thermoresistible__bu ---------MWTAADLPSFSDRTVIVTGANSGLGLVTARELARVGARVII
Mflv_1940|M.gilvum_PYR-GCK -------MSDWTAADLPSFAGRTVIVTGANSGLGLVTARELARVGATTIL
Mvan_4786|M.vanbaalenii_PYR-1 -------MSDWTAADLPSFAGRTVIVTGANSGLGLITARELARVGARTIL
MAB_1144c|M.abscessus_ATCC_199 ---MSKSTATWSAAKLPSFEGRTVIITGANSGLGLETARELARVGAHVIM
Mb0447c|M.bovis_AF2122/97 MTANDNKTRKWSAADVPDQSGRVVVVTGANTGIGYHTAAVFADRGAHVVL
Rv0439c|M.tuberculosis_H37Rv MTANDNKTRKWSAADVPDQSGRVVVVTGANTGIGYHTAAVFADRGAHVVL
MMAR_0757|M.marinum_M MSANGKAKSHWSASDIPDQSGRVVVVTGANTGLGYHTAEALAGRGAHVVL
MUL_1391|M.ulcerans_Agy99 MSAKGKAKSHWSASDIPDQSGRVVVVTGANTGLGYHTAEALADRGAHVVL
MLBr_00315|M.leprae_Br4923 -------MAKWTTADIPDQTGRVAVITGANTGLGYQTALALAEHGAHVVL
MAV_2670|M.avium_104 ---------------------MRIIVTGGNSGVGKATAHAMAAAGHEVVI
::**.*:*:* ** :* * .::
MSMEG_5430|M.smegmatis_MC2_155 AVRNLDKGTAAAETMPG----GIVEVRKLDLQDLASVHEFADTVE----T
TH_3199|M.thermoresistible__bu AVRNTTKGEAAAEQMTGPG-HGPVEVRALDLQNLASVREFAAGVEG---D
Mflv_1940|M.gilvum_PYR-GCK AVRNLDKGRAAADSMSG-----DVEVRRLDLQDLSSVREFADGVD----S
Mvan_4786|M.vanbaalenii_PYR-1 AVRNLDKGNTAAATMTG-----DVEVRSLDLQNLASVRAFADGVD----S
MAB_1144c|M.abscessus_ATCC_199 AVRNTEKGNAAQASIKG-----STEVRQVDVADLASVRAFADTVE----G
Mb0447c|M.bovis_AF2122/97 AVRNLEKGNAARARIMAARPGAHVTLQQLDLCSLDSVRAAADALRTAYPR
Rv0439c|M.tuberculosis_H37Rv AVRNLEKGNAARARIMAARPGAHVTLQQLDLCSLDSVRAAADALRTAYPR
MMAR_0757|M.marinum_M AVRNPEKGNAAVAQIVAAKPQADVTLQALDLSSLDSVRSAADALRSAYPR
MUL_1391|M.ulcerans_Agy99 AVRNPEKGNAAVAQIVAAKPQADVTLQALDLSSLDSVRSAADALRSAYPR
MLBr_00315|M.leprae_Br4923 AVRNLDKGKDAAARITATSAQNNVALQELDLASLESVRAAAKQLRSDYDH
MAV_2670|M.avium_104 ACRSLTKAHQAAASMPG-----DVTVRELDLADLTSVRKFADTVD----Y
* *. *. * : . . :: :*: .* **: * :
MSMEG_5430|M.smegmatis_MC2_155 VDVLVNNAGIMAVPLARTVDGFESQIGTNHLGHFALTNLLLPKIS----D
TH_3199|M.thermoresistible__bu VDVLVNNAGIMAVPLARTVDGFESQIGTNHLGHFALTNLLLPQIT----D
Mflv_1940|M.gilvum_PYR-GCK VDVLVNNAGIMAVPYALTADGFESQIGTNHLGHFALTNLLLPKIS----D
Mvan_4786|M.vanbaalenii_PYR-1 VDVLVNNAGIMAVPYAQTVDGFESQIGTNHLGHFALTNLLLPKIS----D
MAB_1144c|M.abscessus_ATCC_199 ADILVNNAGIMMVPPAKTVDGFESQIGTNHLGAFALTNLLLPKLT----D
Mb0447c|M.bovis_AF2122/97 IDVLINNAGVMWTPKQVTKDGFELQFGTNHLGHFALTGLVLDHMLPVPGS
Rv0439c|M.tuberculosis_H37Rv IDVLINNAGVMWTPKQVTKDGFELQFGTNHLGHFALTGLVLDHMLPVPGS
MMAR_0757|M.marinum_M IDLLINNAGVMWTPKQVTKDGFEMQFGTNHLGHFALTGLLLDHLLPVPGS
MUL_1391|M.ulcerans_Agy99 IDLLINNAGVMWTPKQVTKDGFEMQFGTNHLGHFALTGLLLDHLLPVPGS
MLBr_00315|M.leprae_Br4923 IDLLINNAGVMWTPKSTTKDGFELQFGTNHLGHFAFTGLLLDRLLPIVGS
MAV_2670|M.avium_104 VDVLVNNAGVLGLPLTRTADGFEAHMGINHLGHFALTCLLGDRIR----D
*:*:****:: * * **** ::* **** **:* *: :: .
MSMEG_5430|M.smegmatis_MC2_155 RVVTVSSMMH-WIGRINLRDLNWKARPYSAWLAYGQSKLANLMFTSELQR
TH_3199|M.thermoresistible__bu RVVTVSSLMH-WFGYLSLNDLNWKARPYSAWLAYAQSKLANLLFTSELQR
Mflv_1940|M.gilvum_PYR-GCK RVVTVSSMMH-LFGRINLNDLNWKSRPYLAWPAYGQSKLANLLFTSELQR
Mvan_4786|M.vanbaalenii_PYR-1 RVVTVSSMMH-LMGRVNLKDLNWKSRPYLAWPAYGQSKLANLLFTGELQR
MAB_1144c|M.abscessus_ATCC_199 RVIAVSSVAH-RAGKIDLTDLNYERRRYTRAGAYGASKLANLLFTKELQR
Mb0447c|M.bovis_AF2122/97 RVVTVSSQGHRIHAAIHFDDLQWERR-YNRVAAYGQAKLANLLFTYELQR
Rv0439c|M.tuberculosis_H37Rv RVVTVSSQGHRIHAAIHFDDLQWERR-YNRVAAYGQAKLANLLFTYELQR
MMAR_0757|M.marinum_M RVITVSSLGHRIRAAIHFDDLQWERS-YNRVAAYGQSKLANLLFTYELQR
MUL_1391|M.ulcerans_Agy99 RVITVSSLGHRIRAAIHFDDLQWERS-YNRVAAYGQSKLANLLFTYELQR
MLBr_00315|M.leprae_Br4923 RVITVSSLSHRLFADIHFNDLQWECN-YNRVAAYGQSKLANLLFTYELQR
MAV_2670|M.avium_104 RVVLVASTNY-NTARMHFDDLNYHHRRYNPWSAYGESKLANLLFVRELVA
**: *:* : . : : **::. * **. :*****:*. **
MSMEG_5430|M.smegmatis_MC2_155 RLV-ASGSSVRALAAHPGYSATNLQGHSGNRFGTAAAALGNRHLATDVTF
TH_3199|M.thermoresistible__bu RLD-TVGSPVRAVAAHPGYSSTNLQGHTGRRFGDALARIGNRYLATDAEF
Mflv_1940|M.gilvum_PYR-GCK RLS-RAGSPVRAVAAHPGYSATNLQGHSGNPLGEKIARAGNR-MATDADF
Mvan_4786|M.vanbaalenii_PYR-1 RLA-AAGSPVRAVAAHPGYSATNLQGHSGNPLGEKIARAGNR-IATDADF
MAB_1144c|M.abscessus_ATCC_199 RLS-AAGSPVRALTVHPGVAATELQSHSGNPIFELALKTGNL-FAQNAAH
Mb0447c|M.bovis_AF2122/97 RLG-EAGKSTIAVAAHPGGSNTELTRNLPRLIRPVATVLGPL-LFQSPEM
Rv0439c|M.tuberculosis_H37Rv RLG-EAGKSTIAVAAHPGGSNTELTRNLPRLIRPVATVLGPL-LFQSPEM
MMAR_0757|M.marinum_M RLAADSQAATIAVAAHPGGSNTELARNLPRMLVPLANILGPA-LFQSAQM
MUL_1391|M.ulcerans_Agy99 RLAADSQAATIAVAAHPGDSNTELARNLPRMLVPLANILGPA-LFQSAQM
MLBr_00315|M.leprae_Br4923 RLA--TRQTTIAVAAHPGGSRTELTRTLPALIAPIFSVAELF-LTQDAAT
MAV_2670|M.avium_104 RGK-------TAYASDPGMTDTGITRNGSGVLQWAGRVLSPH-IAQSPAD
* * : .** : * : : : .
MSMEG_5430|M.smegmatis_MC2_155 GARQTLYAVSQDLP-GDTFVGPR--FGQWGPTGSVGRSPLARRTDTQRAL
TH_3199|M.thermoresistible__bu GARQTLYAVARDVP-GDSFIGPR--FAMHGPTEQVGRSPLARSAGTARAL
Mflv_1940|M.gilvum_PYR-GCK GARQTLYAVARDVA-GDSFIGPR--FTMFGPTVTTWRSPLARNRTTAAQL
Mvan_4786|M.vanbaalenii_PYR-1 GARQTLYAVAADIP-GDSFIGPR--FSMFGPTVATWRSPLARDRGKAAGL
MAB_1144c|M.abscessus_ATCC_199 GALPTLYAASQDLP-GDTYIGPDGPFEAKGYPTRVGRSGRAQDTETAKGL
Mb0447c|M.bovis_AF2122/97 GALPTLRAATDPTTQGGQYYGPDGFGEQRGHPKVVQSSAQSHDKDLQRRL
Rv0439c|M.tuberculosis_H37Rv GALPTLRAATDPTTQGGQYYGPDGFGEQRGHPKVVQSSAQSHDKDLQRRL
MMAR_0757|M.marinum_M GALPTLRAATDPSVAGGQYYGPDGFAEQRGHPKIVQSSAQSHDEDLQRRL
MUL_1391|M.ulcerans_Agy99 GALPTLRTATDPSAAGGQYYGPDGFAEQRGHPKIVQSSAQSHDEDLQRRL
MLBr_00315|M.leprae_Br4923 GALPTLRAATDAAVLGGQYFGPDGFAEIRGHPKVVASNGKSHDVDRQLRL
MAV_2670|M.avium_104 GAR---------------------------------SEELSRPVD-----
** . ::
MSMEG_5430|M.smegmatis_MC2_155 WELSEQLTGTTYPL-
TH_3199|M.thermoresistible__bu WELSEQLTDTKFPL-
Mflv_1940|M.gilvum_PYR-GCK WTLSEQLTGVEFPL-
Mvan_4786|M.vanbaalenii_PYR-1 WTLSEQLTGTEFPL-
MAB_1144c|M.abscessus_ATCC_199 WRLSEQLTGVSFPL-
Mb0447c|M.bovis_AF2122/97 WTVSEELTGVSFGV-
Rv0439c|M.tuberculosis_H37Rv WTVSEELTGVSFGV-
MMAR_0757|M.marinum_M WTVSEELTGVSFPV-
MUL_1391|M.ulcerans_Agy99 WTVSEELTGVSFPV-
MLBr_00315|M.leprae_Br4923 WAVSEELTGVVYPVG
MAV_2670|M.avium_104 WGVSGDLR---FG--
* :* :* :