For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SKSTATWSAAKLPSFEGRTVIITGANSGLGLETARELARVGAHVIMAVRNTEKGNAAQASIKGSTEVRQV DVADLASVRAFADTVEGADILVNNAGIMMVPPAKTVDGFESQIGTNHLGAFALTNLLLPKLTDRVIAVSS VAHRAGKIDLTDLNYERRRYTRAGAYGASKLANLLFTKELQRRLSAAGSPVRALTVHPGVAATELQSHSG NPIFELALKTGNLFAQNAAHGALPTLYAASQDLPGDTYIGPDGPFEAKGYPTRVGRSGRAQDTETAKGLW RLSEQLTGVSFPL*
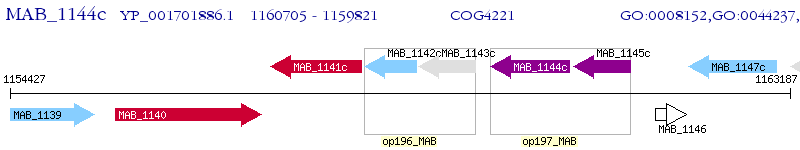
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1144c | - | - | 100% (294) | short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_0646c | - | 5e-62 | 45.95% (309) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0643 | - | 1e-61 | 46.75% (308) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0069 | - | 3e-66 | 48.68% (302) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1940 | - | 6e-95 | 63.07% (287) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0068 | - | 7e-66 | 48.34% (302) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 1e-60 | 46.43% (308) | short chain dehydrogenase |
| M. marinum M | MMAR_0757 | - | 2e-65 | 48.05% (308) | dehydrogenase/reductase |
| M. avium 104 | MAV_1153 | - | 6e-92 | 62.50% (288) | retinol dehydrogenase 13 |
| M. smegmatis MC2 155 | MSMEG_5430 | - | 9e-94 | 63.32% (289) | retinol dehydrogenase 13 |
| M. thermoresistible (build 8) | TH_3199 | - | 1e-87 | 59.73% (293) | PUTATIVE retinol dehydrogenase 13 |
| M. ulcerans Agy99 | MUL_1391 | - | 7e-65 | 47.73% (308) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4786 | - | 2e-97 | 63.76% (287) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5430|M.smegmatis_MC2_155 -------MTQWTSADLPSFRGRTVVITGANSGLGLVTARELARVGAKVIV
TH_3199|M.thermoresistible__bu ---------MWTAADLPSFSDRTVIVTGANSGLGLVTARELARVGARVII
Mflv_1940|M.gilvum_PYR-GCK -------MSDWTAADLPSFAGRTVIVTGANSGLGLVTARELARVGATTIL
Mvan_4786|M.vanbaalenii_PYR-1 -------MSDWTAADLPSFAGRTVIVTGANSGLGLITARELARVGARTIL
MAV_1153|M.avium_104 -------MTGWTAADLPSFAQRTVVITGANSGLGAVTARELARRGATVIM
MAB_1144c|M.abscessus_ATCC_199 ---MSKSTATWSAAKLPSFEGRTVIITGANSGLGLETARELARVGAHVIM
Mb0069|M.bovis_AF2122/97 -------MTKWTAADIPDQTGRTAVITGANTGLGFETAAALAAHGAHVVL
Rv0068|M.tuberculosis_H37Rv -------MTKWTAADIPDQTGRTAVITGANTGLGFETAAALAAHGAHVVL
MLBr_00315|M.leprae_Br4923 -------MAKWTTADIPDQTGRVAVITGANTGLGYQTALALAEHGAHVVL
MMAR_0757|M.marinum_M MSANGKAKSHWSASDIPDQSGRVVVVTGANTGLGYHTAEALAGRGAHVVL
MUL_1391|M.ulcerans_Agy99 MSAKGKAKSHWSASDIPDQSGRVVVVTGANTGLGYHTAEALADRGAHVVL
*:::.:*. *..::****:*** ** ** ** .::
MSMEG_5430|M.smegmatis_MC2_155 AVRNLDKGTAAAETMPG----GIVEVRKLDLQDLASVHEFADTVE----T
TH_3199|M.thermoresistible__bu AVRNTTKGEAAAEQMTGPG-HGPVEVRALDLQNLASVREFAAGVEG---D
Mflv_1940|M.gilvum_PYR-GCK AVRNLDKGRAAADSMSG-----DVEVRRLDLQDLSSVREFADGVD----S
Mvan_4786|M.vanbaalenii_PYR-1 AVRNLDKGNTAAATMTG-----DVEVRSLDLQNLASVRAFADGVD----S
MAV_1153|M.avium_104 AVRDTRKGEAAARTMAG-----QVEVRELDLQDLSSVRRFADGVS----G
MAB_1144c|M.abscessus_ATCC_199 AVRNTEKGNAAQASIKG-----STEVRQVDVADLASVRAFADTVE----G
Mb0069|M.bovis_AF2122/97 AVRNLDKGKQAAARITEATPGAEVELQELDLTSLASVRAAAAQLKSDHQR
Rv0068|M.tuberculosis_H37Rv AVRNLDKGKQAAARITEATPGAEVELQELDLTSLASVRAAAAQLKSDHQR
MLBr_00315|M.leprae_Br4923 AVRNLDKGKDAAARITATSAQNNVALQELDLASLESVRAAAKQLRSDYDH
MMAR_0757|M.marinum_M AVRNPEKGNAAVAQIVAAKPQADVTLQALDLSSLDSVRSAADALRSAYPR
MUL_1391|M.ulcerans_Agy99 AVRNPEKGNAAVAQIVAAKPQADVTLQALDLSSLDSVRSAADALRSAYPR
***: ** * : . :: :*: .* **: * :
MSMEG_5430|M.smegmatis_MC2_155 VDVLVNNAGIMAVPLARTVDGFESQIGTNHLGHFALTNLLLPKIS----D
TH_3199|M.thermoresistible__bu VDVLVNNAGIMAVPLARTVDGFESQIGTNHLGHFALTNLLLPQIT----D
Mflv_1940|M.gilvum_PYR-GCK VDVLVNNAGIMAVPYALTADGFESQIGTNHLGHFALTNLLLPKIS----D
Mvan_4786|M.vanbaalenii_PYR-1 VDVLVNNAGIMAVPYAQTVDGFESQIGTNHLGHFALTNLLLPKIS----D
MAV_1153|M.avium_104 ADVLINNAGIMAVPYALTVDGFESQIGTNHLGHFALTNLLLPRLT----D
MAB_1144c|M.abscessus_ATCC_199 ADILVNNAGIMMVPPAKTVDGFESQIGTNHLGAFALTNLLLPKLT----D
Mb0069|M.bovis_AF2122/97 IDLLINNAGVMYTPRQTTADGFEMQFGTNHLGHFALTGLLIDRLLPVAGS
Rv0068|M.tuberculosis_H37Rv IDLLINNAGVMYTPRQTTADGFEMQFGTNHLGHFALTGLLIDRLLPVAGS
MLBr_00315|M.leprae_Br4923 IDLLINNAGVMWTPKSTTKDGFELQFGTNHLGHFAFTGLLLDRLLPIVGS
MMAR_0757|M.marinum_M IDLLINNAGVMWTPKQVTKDGFEMQFGTNHLGHFALTGLLLDHLLPVPGS
MUL_1391|M.ulcerans_Agy99 IDLLINNAGVMWTPKQVTKDGFEMQFGTNHLGHFALTGLLLDHLLPVPGS
*:*:****:* .* * **** *:****** **:*.**: :: .
MSMEG_5430|M.smegmatis_MC2_155 RVVTVSSMMHWI-GRINLRDLNWKARPYSAWLAYGQSKLANLMFTSELQR
TH_3199|M.thermoresistible__bu RVVTVSSLMHWF-GYLSLNDLNWKARPYSAWLAYAQSKLANLLFTSELQR
Mflv_1940|M.gilvum_PYR-GCK RVVTVSSMMHLF-GRINLNDLNWKSRPYLAWPAYGQSKLANLLFTSELQR
Mvan_4786|M.vanbaalenii_PYR-1 RVVTVSSMMHLM-GRVNLKDLNWKSRPYLAWPAYGQSKLANLLFTGELQR
MAV_1153|M.avium_104 RVVTVSSMAHWP-GRINLEDLNWRSRRYSPWLAYSQSKLANLLFTSELQR
MAB_1144c|M.abscessus_ATCC_199 RVIAVSSVAHRA-GKIDLTDLNYERRRYTRAGAYGASKLANLLFTKELQR
Mb0069|M.bovis_AF2122/97 RVVTISSVGHRIRAAIHFDDLQWERR-YRRVAAYGQAKLANLLFTYELQR
Rv0068|M.tuberculosis_H37Rv RVVTISSVGHRIRAAIHFDDLQWERR-YRRVAAYGQAKLANLLFTYELQR
MLBr_00315|M.leprae_Br4923 RVITVSSLSHRLFADIHFNDLQWECN-YNRVAAYGQSKLANLLFTYELQR
MMAR_0757|M.marinum_M RVITVSSLGHRIRAAIHFDDLQWERS-YNRVAAYGQSKLANLLFTYELQR
MUL_1391|M.ulcerans_Agy99 RVITVSSLGHRIRAAIHFDDLQWERS-YNRVAAYGQSKLANLLFTYELQR
**:::**: * . : : **::. * **. :*****:** ****
MSMEG_5430|M.smegmatis_MC2_155 RLVASG-SSVRALAAHPGYSATNLQGHSGNRFGTAAAALGNRHLATDVTF
TH_3199|M.thermoresistible__bu RLDTVG-SPVRAVAAHPGYSSTNLQGHTGRRFGDALARIGNRYLATDAEF
Mflv_1940|M.gilvum_PYR-GCK RLSRAG-SPVRAVAAHPGYSATNLQGHSGNPLGEKIARAGNR-MATDADF
Mvan_4786|M.vanbaalenii_PYR-1 RLAAAG-SPVRAVAAHPGYSATNLQGHSGNPLGEKIARAGNR-IATDADF
MAV_1153|M.avium_104 RLTAAG-SPLRALAAHPGYSHTNLQGASGRKLGDALMSAATRVVATDADF
MAB_1144c|M.abscessus_ATCC_199 RLSAAG-SPVRALTVHPGVAATELQSHSGNPIFELALKTGNL-FAQNAAH
Mb0069|M.bovis_AF2122/97 RLAPG--GTTIAVASHPGVSNTELVRNMPRPLVAVAAILAP--LMQDAEL
Rv0068|M.tuberculosis_H37Rv RLAPG--GTTIAVASHPGVSNTEVVRNMPRPLVAVAAILAP--LMQDAEL
MLBr_00315|M.leprae_Br4923 RLATR--QTTIAVAAHPGGSRTELTRTLPALIAPIFSVAELF-LTQDAAT
MMAR_0757|M.marinum_M RLAADSQAATIAVAAHPGGSNTELARNLPRMLVPLANILGPA-LFQSAQM
MUL_1391|M.ulcerans_Agy99 RLAADSQAATIAVAAHPGDSNTELARNLPRMLVPLANILGPA-LFQSAQM
** . *:: *** : *:: : . ..
MSMEG_5430|M.smegmatis_MC2_155 GARQTLYAVSQ-DLPGDTFVGPR--FGQWGPTGSVGRSPLARRTDTQRAL
TH_3199|M.thermoresistible__bu GARQTLYAVAR-DVPGDSFIGPR--FAMHGPTEQVGRSPLARSAGTARAL
Mflv_1940|M.gilvum_PYR-GCK GARQTLYAVAR-DVAGDSFIGPR--FTMFGPTVTTWRSPLARNRTTAAQL
Mvan_4786|M.vanbaalenii_PYR-1 GARQTLYAVAA-DIPGDSFIGPR--FSMFGPTVATWRSPLARDRGKAAGL
MAV_1153|M.avium_104 GARQTLYAASQ-DLTGDTFVGPR--FGYLGRTQPVGRSRRAKDAGMAAAL
MAB_1144c|M.abscessus_ATCC_199 GALPTLYAASQ-DLPGDTYIGPDGPFEAKGYPTRVGRSGRAQDTETAKGL
Mb0069|M.bovis_AF2122/97 GALPTLRAATDPAVRGGQYFGPDGFGEIRGYPKVVASSAQSHDEQLQRRL
Rv0068|M.tuberculosis_H37Rv GALPTLRAATDPAVRGGQYFGPDGFGEIRGYPKVVASSAQSHDEQLQRRL
MLBr_00315|M.leprae_Br4923 GALPTLRAATDAAVLGGQYFGPDGFAEIRGHPKVVASNGKSHDVDRQLRL
MMAR_0757|M.marinum_M GALPTLRAATDPSVAGGQYYGPDGFAEQRGHPKIVQSSAQSHDEDLQRRL
MUL_1391|M.ulcerans_Agy99 GALPTLRTATDPSAAGGQYYGPDGFAEQRGHPKIVQSSAQSHDEDLQRRL
** ** :.: *. : ** * . . . :: *
MSMEG_5430|M.smegmatis_MC2_155 WELSEQLTGTTYPL-
TH_3199|M.thermoresistible__bu WELSEQLTDTKFPL-
Mflv_1940|M.gilvum_PYR-GCK WTLSEQLTGVEFPL-
Mvan_4786|M.vanbaalenii_PYR-1 WTLSEQLTGTEFPL-
MAV_1153|M.avium_104 WALSEQLTKTEFPL-
MAB_1144c|M.abscessus_ATCC_199 WRLSEQLTGVSFPL-
Mb0069|M.bovis_AF2122/97 WAVSEELTGVVYPVG
Rv0068|M.tuberculosis_H37Rv WAVSEELTGVVYPVG
MLBr_00315|M.leprae_Br4923 WAVSEELTGVVYPVG
MMAR_0757|M.marinum_M WTVSEELTGVSFPV-
MUL_1391|M.ulcerans_Agy99 WTVSEELTGVSFPV-
* :**:** . :*: