For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRMAEEGADIIAVDLAGPLPDSVRYPSATADDFAETVEMVKATGGRIMATAADIRDLDALRTAVDEGVEA YGRLDVIVANAGICVPAPWNEITPESFRDIIDVNVTGTWNTVMAGAQRIIDGGRGGSIILISSYAGVKVQ PYMVHYTASKHAVTGMARAFAAELGKHDIRVNSVHPGPVNTPMGGGDMIAALFAAIETNPTLNNMGTPFL PQWACEPEDVAAAVCWLASDESRYVTATRLSVDQGMAQF
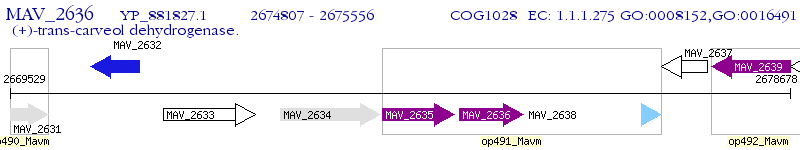
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2636 | - | - | 100% (249) | carveol dehydrogenase |
| M. avium 104 | MAV_1570 | - | e-100 | 68.67% (249) | carveol dehydrogenase |
| M. avium 104 | MAV_5244 | - | 3e-88 | 65.31% (245) | cholesterol oxidase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0706 | fabG | 6e-53 | 41.90% (253) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4371 | - | 1e-105 | 73.49% (249) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0687 | fabG | 7e-53 | 41.90% (253) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 6e-21 | 32.99% (194) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0073c | - | 1e-49 | 45.06% (253) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 3e-56 | 47.01% (251) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2204 | - | 1e-103 | 73.90% (249) | carveol dehydrogenase ((+)-trans-carveol dehydrogenase) |
| M. thermoresistible (build 8) | TH_3169 | - | 8e-96 | 68.67% (249) | PUTATIVE carveol dehydrogenase |
| M. ulcerans Agy99 | MUL_0768 | - | 4e-51 | 41.83% (251) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_1973 | - | 1e-101 | 71.49% (249) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0706|M.bovis_AF2122/97 -MSARGGSLHGRVAFVTGAARAQGRSHAVRLAREGADIVALDICAPVSGS
Rv0687|M.tuberculosis_H37Rv -MSARGGSLHGRVAFVTGAARAQGRSHAVRLAREGADIVALDICAPVSGS
MUL_0768|M.ulcerans_Agy99 ----------------------------MRLAREGADIIAMDICGPVSES
Mflv_4371|M.gilvum_PYR-GCK --MRLE----GRVAFITGAARGQGRAHAVRMAKEGADIIAVDIAGPLPPC
Mvan_1973|M.vanbaalenii_PYR-1 -MSRLE----GRVAFITGAARGQGRAHAVRMATEGADIIAVDIAGPLPPC
MSMEG_2204|M.smegmatis_MC2_155 MAERLT----GRVAFITGAARGQGRAHAVRMAKEGADIIAVDIAGPLPPS
TH_3169|M.thermoresistible__bu MTGRLA----GRVAFITGAARGQGRAHAVRMAAEGADIIAVDIAGKLPDC
MAV_2636|M.avium_104 ----------------------------MRMAEEGADIIAVDLAGPLPDS
MAB_0073c|M.abscessus_ATCC_199 -MTRLG----GRVAVVTGAGKGMGRTHCERLAEEGSDVIAID--------
MMAR_4754|M.marinum_M MTEQLGGRVAGKVAFITGAARGQGRSHAVRLAQEGADIIAVDVCKPIVEN
MLBr_01807|M.leprae_Br4923 --------------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLA
: * . .::. .
Mb0706|M.bovis_AF2122/97 VTYPPATSEDLGETVRAVEAEGRKVLAREVDIRDDAELRRLVADGVEQFG
Rv0687|M.tuberculosis_H37Rv VTYPPATSEDLGETVRAVEAEGRKVLAREVDIRDDAELRRLVADGVEQFG
MUL_0768|M.ulcerans_Agy99 ITYAPATAADLSETIRAVESEGRKVLARQADVRDSAALQQLVADGVEEFG
Mflv_4371|M.gilvum_PYR-GCK VPYDHATADDLAETVELVKATGRRIIASAVDTRDGDALTAAVDSGVAELG
Mvan_1973|M.vanbaalenii_PYR-1 VPYDHATPDDLAETVKLVEATGRRILASKVDTRDGEALRATVEKGVAELG
MSMEG_2204|M.smegmatis_MC2_155 VPYDSATPEDLAETTRLVEATGRRILAARADTRDLEALRDVVGKGVAEFG
TH_3169|M.thermoresistible__bu VPYEPATPEDLDETVRLVTATGRRIHAAVADTRDLDGLREAVDAGVAALG
MAV_2636|M.avium_104 VRYPSATADDFAETVEMVKATGGRIMATAADIRDLDALRTAVDEGVEAYG
MAB_0073c|M.abscessus_ATCC_199 --LPGS--DDLAQTAAAVEEHGRRCVWAFADVRDAAALTEAIDTGVRMLG
MMAR_4754|M.marinum_M TTIPASTPEDLAETADLVKGHNRRIFTAEADVRDYDALKAAVDAGVDELG
MLBr_01807|M.leprae_Br4923 VARRLVADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQG
. . . * * : . *
Mb0706|M.bovis_AF2122/97 RLDIVVANAGVLGW-GRLWELTDEQWETVIGVNLTGTWRTLRATVPAMID
Rv0687|M.tuberculosis_H37Rv RLDIVVANAGVLGW-GRLWELTDEQWETVIGVNLTGTWRTLRATVPAMID
MUL_0768|M.ulcerans_Agy99 RLDVMVANAGVFGW-GRLWELTDEQWDTVIGVNLSGTWRTLRAAVPAMIE
Mflv_4371|M.gilvum_PYR-GCK RLDIIVANAGVTAP-QAWDDISAEDFRDVIDVNVTGTWNTVMAGAHKIVD
Mvan_1973|M.vanbaalenii_PYR-1 RLDVIVANAGITAP-QAWDDIGADDFRDVMDVNVTGTWNTVMAGAHKIID
MSMEG_2204|M.smegmatis_MC2_155 RLDVIVANAGITVP-EPWNETTPESFRDVMDINVTGTWNTVMAGAQHIID
TH_3169|M.thermoresistible__bu RLDVIVANAGICAP-QTWDAITPADFRDVMDINVTGTWNTVMAGARHIID
MAV_2636|M.avium_104 RLDVIVANAGICVP-APWNEITPESFRDIIDVNVTGTWNTVMAGAQRIID
MAB_0073c|M.abscessus_ATCC_199 RLDIVVANAGVYDGLAPSCELTEDSWRRALDVNLTGAWHTVKAGAAHLLD
MMAR_4754|M.marinum_M RLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAAVPHLIA
MLBr_01807|M.leprae_Br4923 PVEVLVANAGITSD-MLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIK
::::*****: .: :.:*::*.:. ::
Mb0706|M.bovis_AF2122/97 AGNGGSIVVVSSSAGLKATPGNGHYAASKHALVALTNTLAIELGEFGIRV
Rv0687|M.tuberculosis_H37Rv AGNGGSIVVVSSSAGLKATPGNGHYAASKHALVALTNTLAIELGEFGIRV
MUL_0768|M.ulcerans_Agy99 AGNGGSIIVVSSSAGLKATPGNGHYAASKHGLVALTNTLAIELGEYDIGV
Mflv_4371|M.gilvum_PYR-GCK GGRGGSIILISSAAGIKLQPFMIHYTASKHAVTGMARAFAAELGKHSIRV
Mvan_1973|M.vanbaalenii_PYR-1 GGRGGSIILISSAAGIKLQPFMIHYTASKHAVTGMARAFAAELGKHSIRV
MSMEG_2204|M.smegmatis_MC2_155 GGRGGSIILISSAAGIKMQPFMVHYTASKHAVAGMARAFAAELGKHRIRV
TH_3169|M.thermoresistible__bu GRRGGSIILISSLAGMKLQPFMVHYTTSKHAVAGMARAFAAELGRHRIRV
MAV_2636|M.avium_104 GGRGGSIILISSYAGVKVQPYMVHYTASKHAVTGMARAFAAELGKHDIRV
MAB_0073c|M.abscessus_ATCC_199 GG---SVIIISSTNGLKGTANTAHYTASKHAVVGLARTAANELGPQGIRV
MMAR_4754|M.marinum_M GGNGGSIVLTSSVGGMKAYPHCGNYVAAKHGVVGLMRSFAVELGQHMIRV
MLBr_01807|M.leprae_Br4923 QRFG-RYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITA
:. .* *:. . :*.::* .: .: .: * *** * .
Mb0706|M.bovis_AF2122/97 NSIHPYSVDTPMIEPEAMIQTFA---KHPGYVHSFPPMP---LQPKGFMT
Rv0687|M.tuberculosis_H37Rv NSIHPYSVDTPMIEPEAMIQTFA---KHPGYVHSFPPMP---LQPKGFMT
MUL_0768|M.ulcerans_Agy99 NSIHPYSVETPMIEPDVMMQVFG---EHPRFLHSFPPMP---LQYKGLMT
Mflv_4371|M.gilvum_PYR-GCK NSVHPGPVVTDMGTGDMVTALGKAMETNPQLSNMMTP-----FLPTWAVE
Mvan_1973|M.vanbaalenii_PYR-1 NSVHPGPVLTDMGTGDMVTALGKAMETNPQLSNMMTP-----FLPTWAVE
MSMEG_2204|M.smegmatis_MC2_155 NSLHPGAVNTPMGTGDMLAALTRANDTNPGLMQMVTP-----FLPDYIAE
TH_3169|M.thermoresistible__bu NSLHPGAVATDMGGGEMVSALAATGESNPPLTQMITP-----FLPDWIAQ
MAV_2636|M.avium_104 NSVHPGPVNTPMGGGDMIAALFAAIETNPTLNNMGTP-----FLPQWACE
MAB_0073c|M.abscessus_ATCC_199 NTVHPGAVSTGMILNPAVIARLRPDLEHPTVEDAAEALAARTLLPVPWVE
MMAR_4754|M.marinum_M NSVHPTHVRTPMLHNEGTFKMFRPDLENPGPDDMAPICQLFHTLPIPWVE
MLBr_01807|M.leprae_Br4923 NVVAPGFIDTEMTRAMDSKYQEAALQAIPLG---------------RIGA
* : * : * * *
Mb0706|M.bovis_AF2122/97 PDEISDVVVWLAGDGSGALSGNQIPVDKGALKY-
Rv0687|M.tuberculosis_H37Rv PDEISDVVVWLAGDGSGALSGNQIPVDKGALKY-
MUL_0768|M.ulcerans_Agy99 SEEVSDVVVWLAGDGSGTLSGAQIPVDKGALKY-
Mflv_4371|M.gilvum_PYR-GCK PEDIADAVCWLASDESKFVTASAISVDQGSTHY-
Mvan_1973|M.vanbaalenii_PYR-1 PEDIADAVCWLASDESKFVTASAISVDQGSTHY-
MSMEG_2204|M.smegmatis_MC2_155 PEDIADAAVWLASDESRFVTASRVAVDLGSTQF-
TH_3169|M.thermoresistible__bu VDDIADAAVWLASDESRFVTAAKIPVDMGSSQF-
MAV_2636|M.avium_104 PEDVAAAVCWLASDESRYVTATRLSVDQGMAQF-
MAB_0073c|M.abscessus_ATCC_199 AVDISNAVVFLASDEARYITGTQLVVDAGLTQKA
MMAR_4754|M.marinum_M AEDISNAVLFLASDESRYITGVTLPVDAGGCLK-
MLBr_01807|M.leprae_Br4923 VEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH
::: .. :**.: : ::. : ** *