For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPGEGPDLVDKIMASIGEQPTGRPAWMRWLRSHYRRWGLIGVGLFQVAIAAAQISGIDFGMVAGHMHGAM SGEHLMHESTAWLLALGLAMIAAGVWPASASGVAAITGVYSVALLGYVIVDAFDGEVTATRIASHMPLLL GLAFALLVARERVGSRRPGSSDATADAGFAAWAADAPAGRRRGHLRPINRAAPDPTASSTTTRRRIGKPA QLRWLGMIAPSDDEAVTELALSAARGNARALEAFIKATQQDVWRFVAYLCDAGSADDLTQETFLRAIGAI ERFSGRSSARTWLLSIARRVVADHIRHLQSRPRAAVGADPEHVLRTDRHARGFEDLVEVTTMIAGLNPEQ REALLLTQLLGLPYADAAAVCGCPVGTIRSRVARARDALLADGERNDLTG*
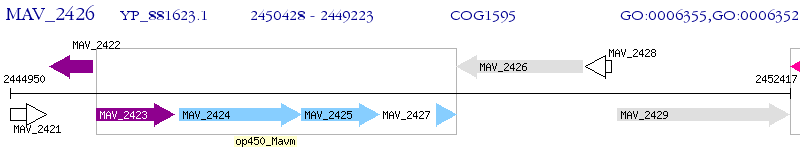
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_2426 | - | - | 100% (401) | RNA polymerase sigma factor SigC |
| M. avium 104 | MAV_4998 | - | 2e-11 | 34.18% (158) | RNA polymerase factor sigma-70 |
| M. avium 104 | MAV_1365 | - | 1e-10 | 29.19% (185) | RNA polymerase sigma-70 factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2095 | sigC | 1e-81 | 85.00% (180) | RNA polymerase sigma factor SigC |
| M. gilvum PYR-GCK | Mflv_2200 | - | 2e-11 | 26.24% (263) | RNA polymerase sigma factor SigE |
| M. tuberculosis H37Rv | Rv2069 | sigC | 1e-81 | 85.00% (180) | RNA polymerase sigma factor SigC |
| M. leprae Br4923 | MLBr_01448 | - | 3e-81 | 83.15% (184) | RNA polymerase sigma factor SigC |
| M. abscessus ATCC 19977 | MAB_3428c | - | 1e-60 | 65.19% (181) | RNA polymerase sigma-C factor |
| M. marinum M | MMAR_3051 | sigC | 1e-80 | 83.70% (184) | RNA polymerase sigma factor, SigC |
| M. smegmatis MC2 155 | MSMEG_5072 | - | 3e-10 | 28.42% (183) | RNA polymerase sigma factor SigE |
| M. thermoresistible (build 8) | TH_1729 | sigC | 1e-58 | 64.80% (179) | PROBABLE RNA POLYMERASE SIGMA FACTOR, ECF SUBFAMILY, SIGC |
| M. ulcerans Agy99 | MUL_2295 | sigC | 5e-81 | 83.33% (186) | RNA polymerase sigma factor SigC |
| M. vanbaalenii PYR-1 | Mvan_4496 | - | 3e-11 | 25.65% (230) | RNA polymerase sigma factor SigE |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_2426|M.avium_104 MTPGEGPDLVDKIMASIGEQPTGRPAWMRWLRSHYRRWGLIGVGLFQVAI
MLBr_01448|M.leprae_Br4923 --------------------------------------------------
Mb2095|M.bovis_AF2122/97 --------------------------------------------------
Rv2069|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3051|M.marinum_M --------------------------------------------------
MUL_2295|M.ulcerans_Agy99 --------------------------------------------------
MAB_3428c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1729|M.thermoresistible__bu --------------------------------------------------
Mflv_2200|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4496|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5072|M.smegmatis_MC2_155 --------------------------------------------------
MAV_2426|M.avium_104 AAAQISGIDFGMVAGHMHGAMSGEHLMHESTAWLLALGLAMIAAGVWPAS
MLBr_01448|M.leprae_Br4923 --------------------------------------------------
Mb2095|M.bovis_AF2122/97 --------------------------------------------------
Rv2069|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3051|M.marinum_M --------------------------------------------------
MUL_2295|M.ulcerans_Agy99 --------------------------------------------------
MAB_3428c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1729|M.thermoresistible__bu --------------------------------------------------
Mflv_2200|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4496|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5072|M.smegmatis_MC2_155 --------------------------------------------------
MAV_2426|M.avium_104 ASGVAAITGVYSVALLGYVIVDAFDGEVTATRIASHMPLLLGLAFALLVA
MLBr_01448|M.leprae_Br4923 --------------------------------------------------
Mb2095|M.bovis_AF2122/97 --------------------------------------------------
Rv2069|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3051|M.marinum_M --------------------------------------------------
MUL_2295|M.ulcerans_Agy99 --------------------------------------------------
MAB_3428c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1729|M.thermoresistible__bu --------------------------------------------------
Mflv_2200|M.gilvum_PYR-GCK ----------------------------------MTEGARTDTVRTARID
Mvan_4496|M.vanbaalenii_PYR-1 ----------------------------------MTDGAFS-----ARPD
MSMEG_5072|M.smegmatis_MC2_155 ----------------------------------MEHDDRR-------AS
MAV_2426|M.avium_104 RERVGSRRPGSSDATADAGFAAWAADAPAGRRRGHLRPINRAAPDPTASS
MLBr_01448|M.leprae_Br4923 --------------------------------------------------
Mb2095|M.bovis_AF2122/97 --------------------------------------------------
Rv2069|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3051|M.marinum_M --------------------------------------------------
MUL_2295|M.ulcerans_Agy99 --------------------------------------------------
MAB_3428c|M.abscessus_ATCC_199 --------------------------------------------------
TH_1729|M.thermoresistible__bu --------------------------------------------------
Mflv_2200|M.gilvum_PYR-GCK SARSRNENASARVAEHD-GSPQGGPHHHLEDPTTTTQAPTSSVSAAAPVA
Mvan_4496|M.vanbaalenii_PYR-1 HARLGNNIPTGRVVAFERSSPSDRTHLETEDPTTTTQAPT---------A
MSMEG_5072|M.smegmatis_MC2_155 QVRAGNNRVVIRVEQIE-----NNAVHEQEGSTTIASQPV---------T
MAV_2426|M.avium_104 TTTRRRIGKPAQLRWLGMIAPSDDEAVTELALSAARGNARAL---EAFIK
MLBr_01448|M.leprae_Br4923 -----------------MIASSGDEAVTDLALAAARGSKRAL---EAFIK
Mb2095|M.bovis_AF2122/97 ----------------MTATASDDEAVTALALSAAKGNGRAL---EAFIK
Rv2069|M.tuberculosis_H37Rv ----------------MTATASDDEAVTALALSAAKGNGRAL---EAFIK
MMAR_3051|M.marinum_M -----------------MTTSPDDEAVTALALAAADGDARAF---EAFIK
MUL_2295|M.ulcerans_Agy99 ---------------MPMTTSPDDEAVTALALAAADGDARAF---EAFIK
MAB_3428c|M.abscessus_ATCC_199 -----------------MATGTDDEQLLELALSAGRGDQSAL---ESLVK
TH_1729|M.thermoresistible__bu ------------------VGLRDDDRVTRLALAAGRGDPAAL---EQFIR
Mflv_2200|M.gilvum_PYR-GCK APVAAPVSMAHREQFTEAEWVEPTDELTGTAVFDATGDKAAMPSWDELVR
Mvan_4496|M.vanbaalenii_PYR-1 APVAAPVTMAHLEQFTDAAWVEPADELTGTAVFDATGDKAAMPSWDELVR
MSMEG_5072|M.smegmatis_MC2_155 ATHAAPVSMAHLEQFTDSDWVEPSDEPTGTAVFDATGDQAAMPSWDELVR
: *: . *. *: : :::
MAV_2426|M.avium_104 ATQQDVWRFVAYLC-DAGSADDLTQETFLRAIGAIERFSGRSSARTWLLS
MLBr_01448|M.leprae_Br4923 ATQQDVWRFVAYLS-DVGNADDLTQETFLRAIGAIERFSGHSSGRTWLLA
Mb2095|M.bovis_AF2122/97 ATQQDVWRFVAYLS-DVGSADDLTQETFLRAIGAIPRFSARSSARTWLLA
Rv2069|M.tuberculosis_H37Rv ATQQDVWRFVAYLS-DVGSADDLTQETFLRAIGAIPRFSARSSARTWLLA
MMAR_3051|M.marinum_M ATQQDVWRFVAYLS-DAGSADDLTQETFLRAIGAIRRFGGRSSARTWLLA
MUL_2295|M.ulcerans_Agy99 ATQQDVWRFVAYLS-DAGSADDLTQETFLRAIGAIRRFGGRSSARTWLLA
MAB_3428c|M.abscessus_ATCC_199 ATQGDVWRFVAYLA-DSGVADDLTQETYLRALGALPRFAGRSTVRTWLLS
TH_1729|M.thermoresistible__bu ATQRDVWRTVAYLA-DPGSADDLTQETFLRAIRSLPKFNGRSSARTWLLS
Mflv_2200|M.gilvum_PYR-GCK QHADRVYRLAYRLSGNQHDAEDLTQETFIRVFRSVQNYQP-GTFEGWLHR
Mvan_4496|M.vanbaalenii_PYR-1 QHADRVYRLAYRLSGNQHDAEDLTQETFIRVFRSVQNYQP-GTFEGWLHR
MSMEG_5072|M.smegmatis_MC2_155 QHADRVYRLAYRLSGNQHDAEDLTQETFIRVFRSVQNYQP-GTFEGWLHR
*:* . *. : *:******::*.: :: .: .: . **
MAV_2426|M.avium_104 IARRVVADHIRHL-QSRPRAAVGADPEHVLRTDRHARGFEDL---VEVTT
MLBr_01448|M.leprae_Br4923 IARRVVADHIRRA-QTRPRTTFDGYPERMLNADRHARGFEDL---VEVTT
Mb2095|M.bovis_AF2122/97 IARHVVADHIRHV-RSRPRTTRGARPEHLIDGDRHARGFEDL---VEVTT
Rv2069|M.tuberculosis_H37Rv IARHVVADHIRHV-RSRPRTTRGARPEHLIDGDRHARGFEDL---VEVTT
MMAR_3051|M.marinum_M IARRVVADHIRHL-QARPRAAHGVDPELLLDRGRRARGFEDL---VEVTT
MUL_2295|M.ulcerans_Agy99 IARRVVADHIRHL-QARPRAAHGVDPELLLDRGRRARGFEDL---VEVTT
MAB_3428c|M.abscessus_ATCC_199 IARRVVADHIRHL-KSRPRIAHGADLEEASLR-RPTHRFEEL---VELKS
TH_1729|M.thermoresistible__bu IARRVVVDQIRHN-QSRPRTSYAVDVHTVLDKRRPDGRFEDI---VEIKL
Mflv_2200|M.gilvum_PYR-GCK ITTNLFLDMVRRRGRIRMEALPEDYDRVPADEPNPEQIYHDSRLGADLQA
Mvan_4496|M.vanbaalenii_PYR-1 ITTNLFLDMVRRRGRIRMEALPEDYDRVPADEPNPEQIYHDSRLGPDLQA
MSMEG_5072|M.smegmatis_MC2_155 ITTNLFLDMVRRRGRIRMEALPEDYDRVPAEDPNPEQIYHDSRLGADLQA
*: .:. * :*: : * . . . :.: ::
MAV_2426|M.avium_104 MIAGLNPEQREALLLTQLLGLPYADAAAVCGCPVGTIRSRVARARDALLA
MLBr_01448|M.leprae_Br4923 MIANLATEQREALLLTQLLGLPYADAAAVCGCPVGTIRSRVARARDALLA
Mb2095|M.bovis_AF2122/97 MIADLTTDQREALLLTQLLGLSYADAAAVCGCPVGTIRSRVARARDALLA
Rv2069|M.tuberculosis_H37Rv MIADLTTDQREALLLTQLLGLSYADAAAVCGCPVGTIRSRVARARDALLA
MMAR_3051|M.marinum_M MIAQLTSDQREALLLTQLLGLSYTDAAAVCGCPVGTIRSRVARARDALLA
MUL_2295|M.ulcerans_Agy99 MIAQLTSDQREALLLTQLLGLSYTDAAAVCGCPVGTIRSRVARARDALLA
MAB_3428c|M.abscessus_ATCC_199 LLAALDDDRREALVLTQVVGLSYAETADVCGCAIGTIRSRVARARDELIG
TH_1729|M.thermoresistible__bu LLDGLDTDRRHALLLTQVLGLSYAEAAEVCGCAVGTIRSRVARARDDLLQ
Mflv_2200|M.gilvum_PYR-GCK ALDSLAPEFRAAVVLCDIEGLSYEEIGATLGVKLGTVRSRIHRGRQALRD
Mvan_4496|M.vanbaalenii_PYR-1 ALDSLAPEFRAAVVLCDIEGLSYEEIGATLGVKLGTVRSRIHRGRQALRE
MSMEG_5072|M.smegmatis_MC2_155 ALDSLPPEFRAAVVLCDIEGLSYEEIGATLGVKLGTVRSRIHRGRQQLRD
: * : * *::* :: **.* : . . * :**:***: *.*: *
MAV_2426|M.avium_104 DGERNDLTG---------
MLBr_01448|M.leprae_Br4923 DTEHNHLTG---------
Mb2095|M.bovis_AF2122/97 DAEPDDLTG---------
Rv2069|M.tuberculosis_H37Rv DAEPDDLTG---------
MMAR_3051|M.marinum_M DNERDDLTG---------
MUL_2295|M.ulcerans_Agy99 DNERDDLTG---------
MAB_3428c|M.abscessus_ATCC_199 WAEPENLTG---------
TH_1729|M.thermoresistible__bu QARPVDRTG---------
Mflv_2200|M.gilvum_PYR-GCK HLARHSDVGAGRDTAKSA
Mvan_4496|M.vanbaalenii_PYR-1 YLARHSDAGAGRDTARSA
MSMEG_5072|M.smegmatis_MC2_155 YLAKHS-----SETAQSA